X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking.
Swale, C., Da Costa, B., Sedano, L., Garzoni, F., McCarthy, A.A., Berger, I., Bieniossek, C., Ruigrok, R.W.H., Delmas, B., Crepin, T.(2020) J Mol Biol 432: 3353-3359
- PubMed: 32222384
- DOI: https://doi.org/10.1016/j.jmb.2020.03.021
- Primary Citation of Related Structures:
6XTE, 6XU2 - PubMed Abstract:
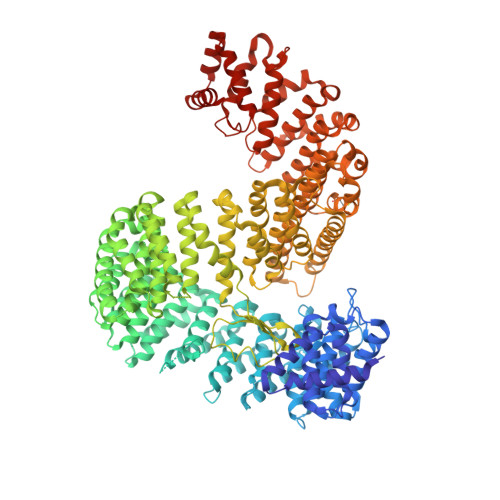
Here, we describe the crystal structures of two distinct isoforms of ligand-free human karyopherin RanBP5 and investigate its global propensity to interact with influenza A virus polymerase. Our results confirm the general architecture and mechanism of the IMB3 karyopherin-β subfamily whilst also highlighting differences with the yeast orthologue Kap121p. Moreover, our results provide insight into the structural flexibility of β-importins in the unbound state. Based on docking of a nuclear localisation sequence, point mutations were designed, which suppress influenza PA-PB1 subcomplex binding to RanBP5 in a binary protein complementation assay.
Organizational Affiliation:
Institut de Biologie Structurale (IBS), University Grenoble Alpes, CEA, CNRS, 38044 Grenoble, France; EMBL Grenoble Outstation, 71 Avenue des Martyrs, BP181, F-38042 Grenoble Cedex 9, France.