Rhs1-CT in complex with cognate immunity protein RhsI1
Hagan, M., Coulthurst, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative deoxyribonuclease RhsA | 155 | Serratia marcescens | Mutation(s): 0 Gene Names: rhsA_2, PWN146_02504 EC: 3.1 | 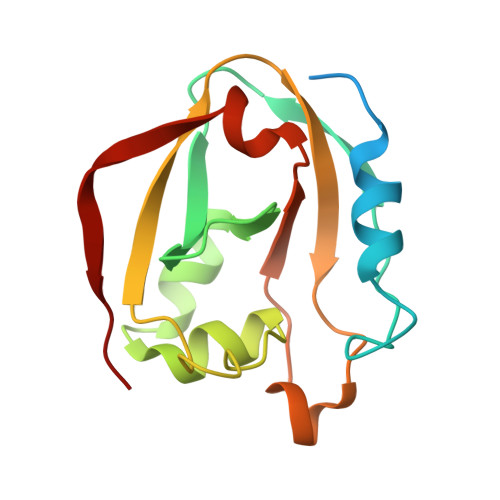 | |
UniProt | |||||
Find proteins for A0A1C3HFI3 (Serratia marcescens) Explore A0A1C3HFI3 Go to UniProtKB: A0A1C3HFI3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1C3HFI3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Secreted protein | 163 | Serratia marcescens | Mutation(s): 0 | 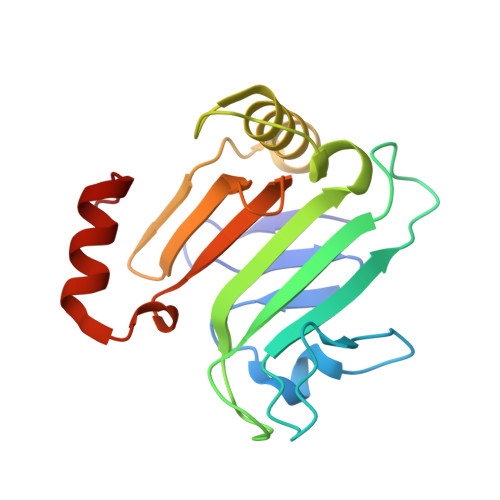 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BTB Query on BTB | E [auth A] | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C8 H19 N O5 OWMVSZAMULFTJU-UHFFFAOYSA-N |  | ||
| BR Query on BR | C [auth A] D [auth A] F [auth B] G [auth B] H [auth B] | BROMIDE ION Br CPELXLSAUQHCOX-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.655 | α = 101.202 |
| b = 44.108 | β = 96.126 |
| c = 46.807 | γ = 114.149 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data reduction |
| Aimless | data scaling |
| CRANK2 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | -- |