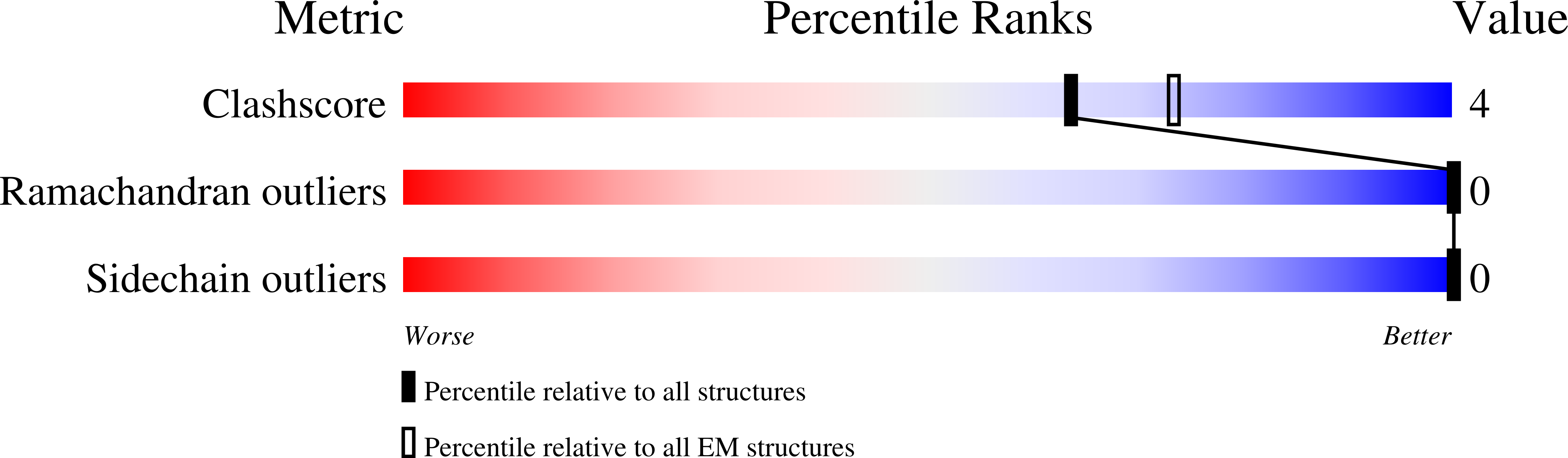Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses.
Moin, S.M., Boyington, J.C., Boyoglu-Barnum, S., Gillespie, R.A., Cerutti, G., Cheung, C.S., Cagigi, A., Gallagher, J.R., Brand, J., Prabhakaran, M., Tsybovsky, Y., Stephens, T., Fisher, B.E., Creanga, A., Ataca, S., Rawi, R., Corbett, K.S., Crank, M.C., Karlsson Hedestam, G.B., Gorman, J., McDermott, A.B., Harris, A.K., Zhou, T., Kwong, P.D., Shapiro, L., Mascola, J.R., Graham, B.S., Kanekiyo, M.(2022) Immunity 55: 2405-2418.e7
- PubMed: 36356572
- DOI: https://doi.org/10.1016/j.immuni.2022.10.015
- Primary Citation of Related Structures:
6XSK - PubMed Abstract:
Current influenza vaccines predominantly induce immunity to the hypervariable hemagglutinin (HA) head, requiring frequent vaccine reformulation. Conversely, the immunosubdominant yet conserved HA stem harbors a supersite that is targeted by broadly neutralizing antibodies (bnAbs), representing a prime target for universal vaccines. Here, we showed that the co-immunization of two HA stem immunogens derived from group 1 and 2 influenza A viruses elicits cross-group protective immunity and neutralizing antibody responses in mice, ferrets, and nonhuman primates (NHPs). Immunized mice were protected from multiple group 1 and 2 viruses, and all animal models showed broad serum-neutralizing activity. A bnAb isolated from an immunized NHP broadly neutralized and protected against diverse viruses, including H5N1 and H7N9. Genetic and structural analyses revealed strong homology between macaque and human bnAbs, illustrating common biophysical constraints for acquiring cross-group specificity. Vaccine elicitation of stem-directed cross-group-protective immunity represents a step toward the development of broadly protective influenza vaccines.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.