Design, Synthesis, and Structure-Activity Relationship Optimization of Pyrazolopyrimidine Amide Inhibitors of Phosphoinositide 3-Kinase gamma (PI3K gamma ).
Mata, G., Miles, D.H., Drew, S.L., Fournier, J., Lawson, K.V., Mailyan, A.K., Sharif, E.U., Yan, X., Beatty, J.W., Banuelos, J., Chen, J., Ginn, E., Chen, A., Gerrick, K.Y., Pham, A.T., Wong, K., Soni, D., Dhanota, P., Shaqfeh, S.G., Meleza, C., Narasappa, N., Singh, H., Zhao, X., Jin, L., Schindler, U., Walters, M.J., Young, S.W., Walker, N.P., Leleti, M.R., Powers, J.P., Jeffrey, J.L.(2022) J Med Chem 65: 1418-1444
- PubMed: 34672584
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01153
- Primary Citation of Related Structures:
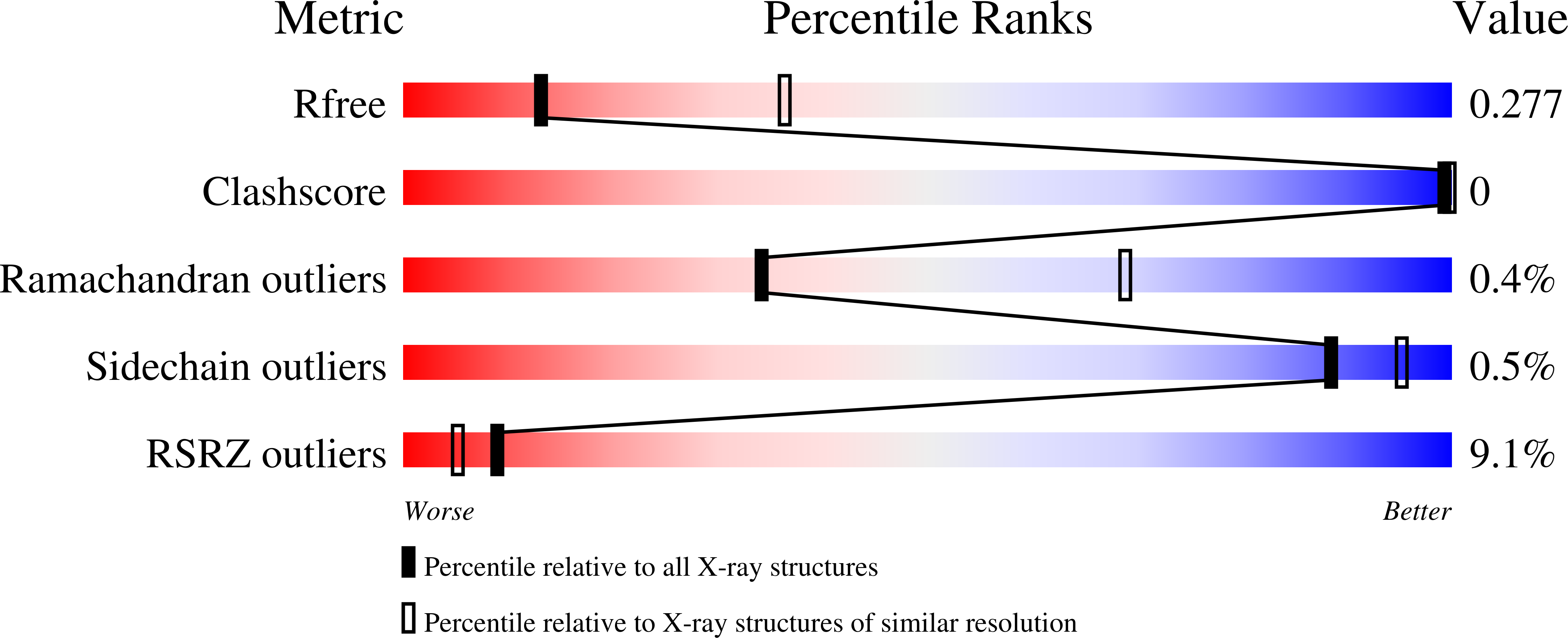
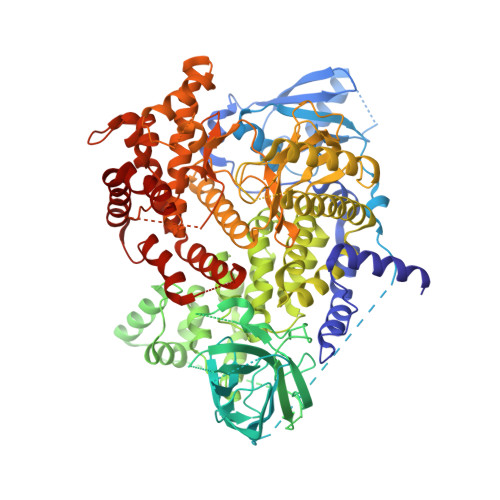
6XRN - PubMed Abstract:
Phosphoinositide-3-kinase γ (PI3Kγ) is highly expressed in immune cells and promotes the production and migration of inflammatory mediators. The inhibition of PI3Kγ has been shown to repolarize the tumor immune microenvironment to a more inflammatory phenotype, thereby controlling immune suppression in cancer. Herein, we report the structure-based optimization of an early lead series of pyrazolopyrimidine isoindolinones, which culminated in the discovery of highly potent and isoform-selective PI3Kγ inhibitors with favorable drug-like properties. X-ray cocrystal structure analysis, molecular docking studies, and detailed structure-activity relationship investigations resulted in the identification of the optimal amide and isoindolinone substituents to achieve a desirable combination of potency, selectivity, and metabolic stability. Preliminary in vitro studies indicate that inhibition of PI3Kγ with compound 56 results in a significant immune response by increasing pro-inflammatory cytokine gene expression in M1 macrophages.
Organizational Affiliation:
Arcus Biosciences, Inc., 3928 Point Eden Way, Hayward, California 94545, United States.