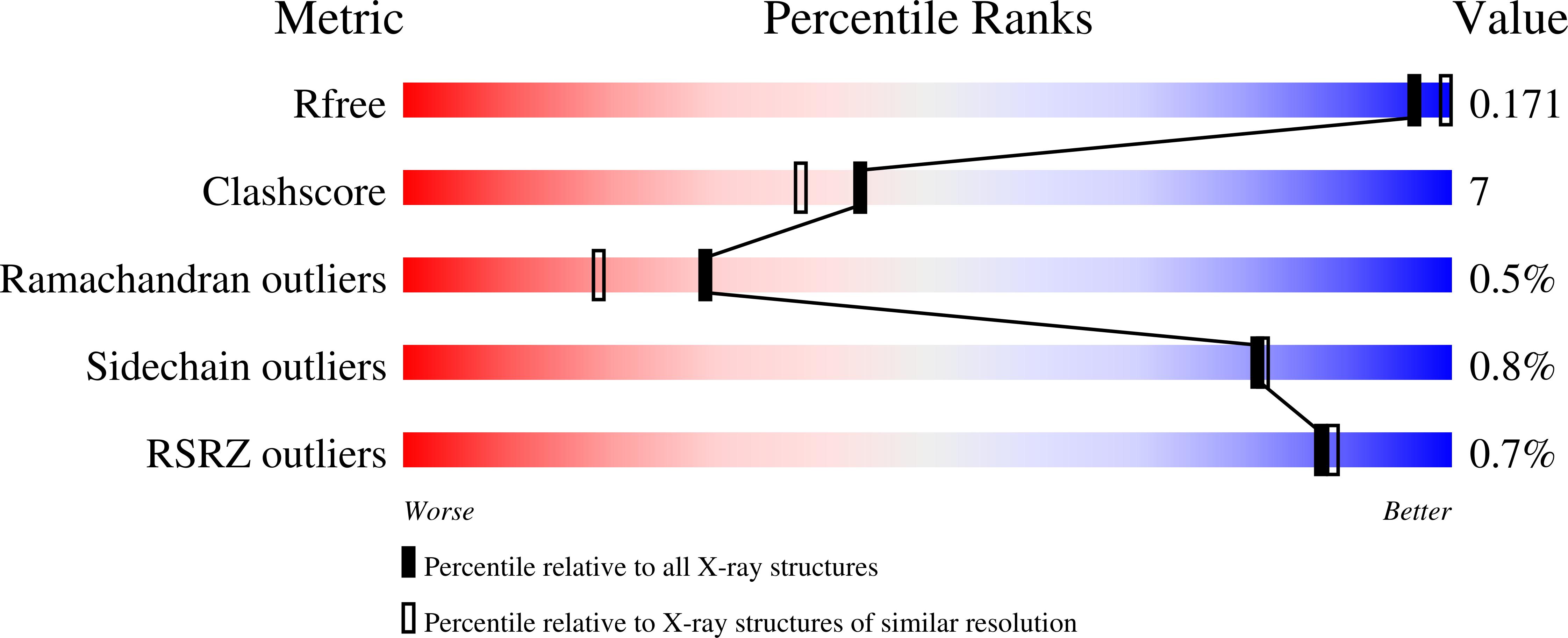
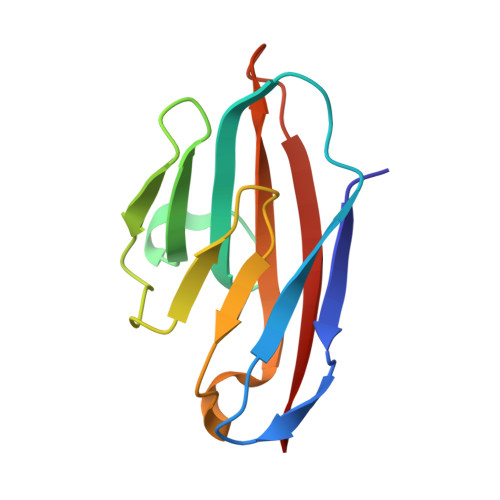
Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Gandhi, A.K., Sun, Z.J., Kim, W.M., Huang, Y.H., Kondo, Y., Bonsor, D.A., Sundberg, E.J., Wagner, G., Kuchroo, V.K., Petsko, G.A., Blumberg, R.S.(2021) Commun Biol 4: 360-360
- PubMed: 33742094
- DOI: https://doi.org/10.1038/s42003-021-01871-2
- Primary Citation of Related Structures:
6XNO, 6XNT, 6XNW, 6XO1 - PubMed Abstract:
Human (h) carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) function depends upon IgV-mediated homodimerization or heterodimerization with host ligands, including hCEACAM5, hTIM-3, PD-1, and a variety of microbial pathogens. However, there is little structural information available on how hCEACAM1 transitions between monomeric and dimeric states which in the latter case is critical for initiating hCEACAM1 activities. We therefore mutated residues within the hCEACAM1 IgV GFCC' face including V39, I91, N97, and E99 and examined hCEACAM1 IgV monomer-homodimer exchange using differential scanning fluorimetry, multi-angle light scattering, X-ray crystallography and/or nuclear magnetic resonance. From these studies, we describe hCEACAM1 homodimeric, monomeric and transition states at atomic resolution and its conformational behavior in solution through NMR assignment of the wildtype (WT) hCEACAM1 IgV dimer and N97A mutant monomer. These studies reveal the flexibility of the GFCC' face and its important role in governing the formation of hCEACAM1 dimers and selective heterodimers.
Organizational Affiliation:
Division of Gastroenterology, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. agandhi2@bwh.harvard.edu.