Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1
Liu, Y.X., Sun, M., Myasnikov, A.G., Elnatan, D., Agard, D.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent molecular chaperone HSC82 | 705 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: HSC82, YMR186W, YM8010.16 | 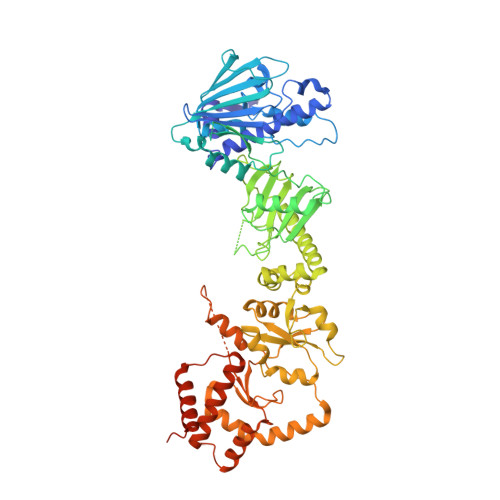 | |
UniProt | |||||
Find proteins for P15108 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P15108 Go to UniProtKB: P15108 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P15108 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hsp90 co-chaperone AHA1 | 350 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: AHA1, YDR214W, YD8142.16, YD8142B.06 | 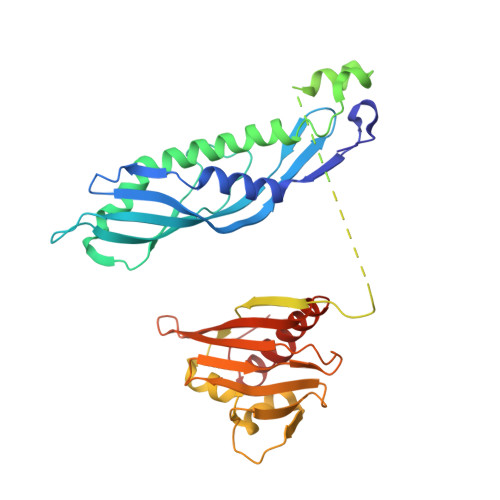 | |
UniProt | |||||
Find proteins for Q12449 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q12449 Go to UniProtKB: Q12449 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12449 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS Query on AGS | E [auth A] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | H [auth B] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| K Query on K | G [auth A], J [auth B] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | F [auth A], I [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Howard Hughes Medical Institute (HHMI) | United States | -- |
| American Heart Association | United States | -- |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | U54CA209891 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | S10OD020054 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | S10OD021741 |