Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
Walker, J.S., Hing, Z.A., Harrington, B., Baumhardt, J., Ozer, H.G., Lehman, A., Giacopelli, B., Beaver, L., Williams, K., Skinner, J.N., Cempre, C.B., Sun, Q., Shacham, S., Stromberg, B.R., Summers, M.K., Abruzzo, L.V., Rassenti, L., Kipps, T.J., Parikh, S., Kay, N.E., Rogers, K.A., Woyach, J.A., Coppola, V., Chook, Y.M., Oakes, C., Byrd, J.C., Lapalombella, R.(2021) J Hematol Oncol 14: 17-17
- PubMed: 33451349
- DOI: https://doi.org/10.1186/s13045-021-01032-2
- Primary Citation of Related Structures:
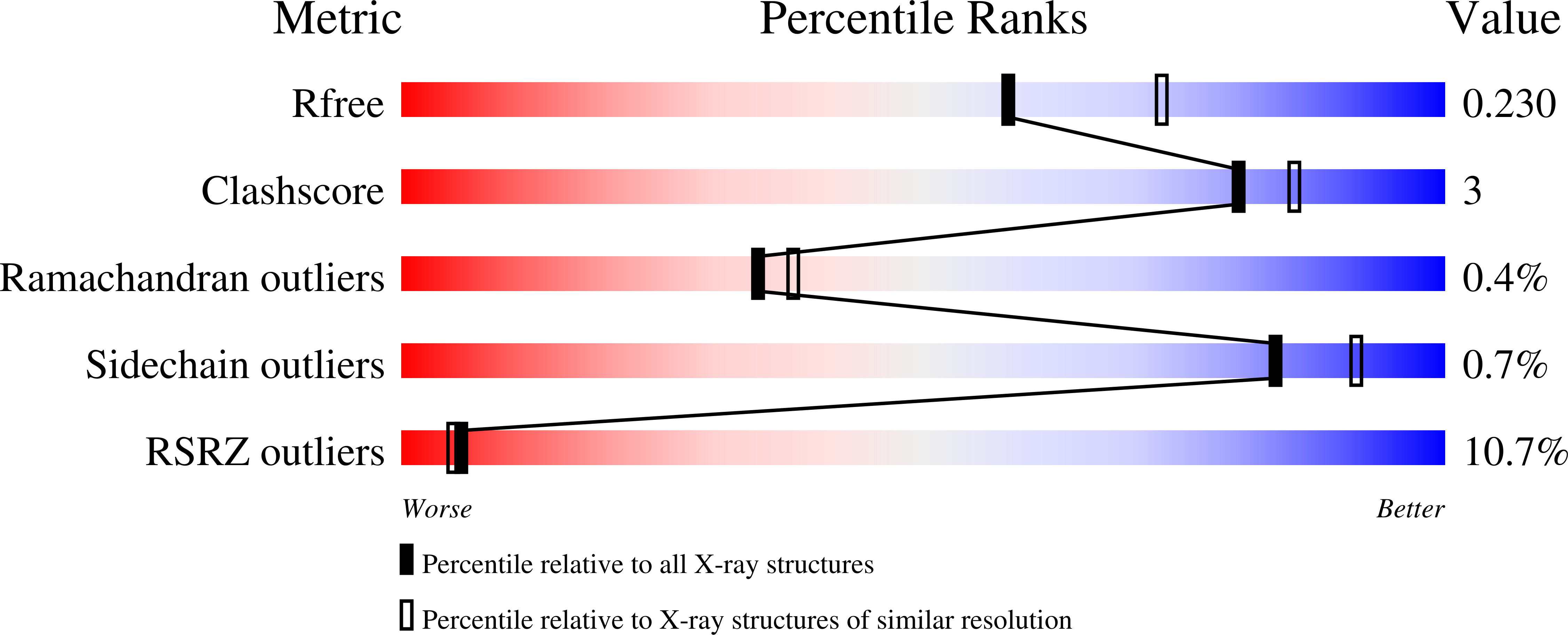
6XJP, 6XJR, 6XJS, 6XJT, 6XJU, 7L5E - PubMed Abstract:
Exportin 1 (XPO1/CRM1) is a key mediator of nuclear export with relevance to multiple cancers, including chronic lymphocytic leukemia (CLL). Whole exome sequencing has identified hot-spot somatic XPO1 point mutations which we found to disrupt highly conserved biophysical interactions in the NES-binding groove, conferring novel cargo-binding abilities and forcing cellular mis-localization of critical regulators. However, the pathogenic role played by change-in-function XPO1 mutations in CLL is not fully understood.
Organizational Affiliation:
Division of Hematology, Department of Internal Medicine, The Ohio State University, 460 OSUCCC, 410 West 12th Avenue, Columbus, OH, 43210, USA.