Structure-guided optimization of a novel class of ASK1 inhibitors with increased sp3character and an exquisite selectivity profile.
Bigi-Botterill, S.V., Ivetac, A., Bradshaw, E.L., Cole, D., Dougan, D.R., Ermolieff, J., Halkowycz, P., Johnson, B., McBride, C., Pickens, J., Sabat, M., Swann, S.(2020) Bioorg Med Chem Lett 30: 127405-127405
- PubMed: 32738982
- DOI: https://doi.org/10.1016/j.bmcl.2020.127405
- Primary Citation of Related Structures:
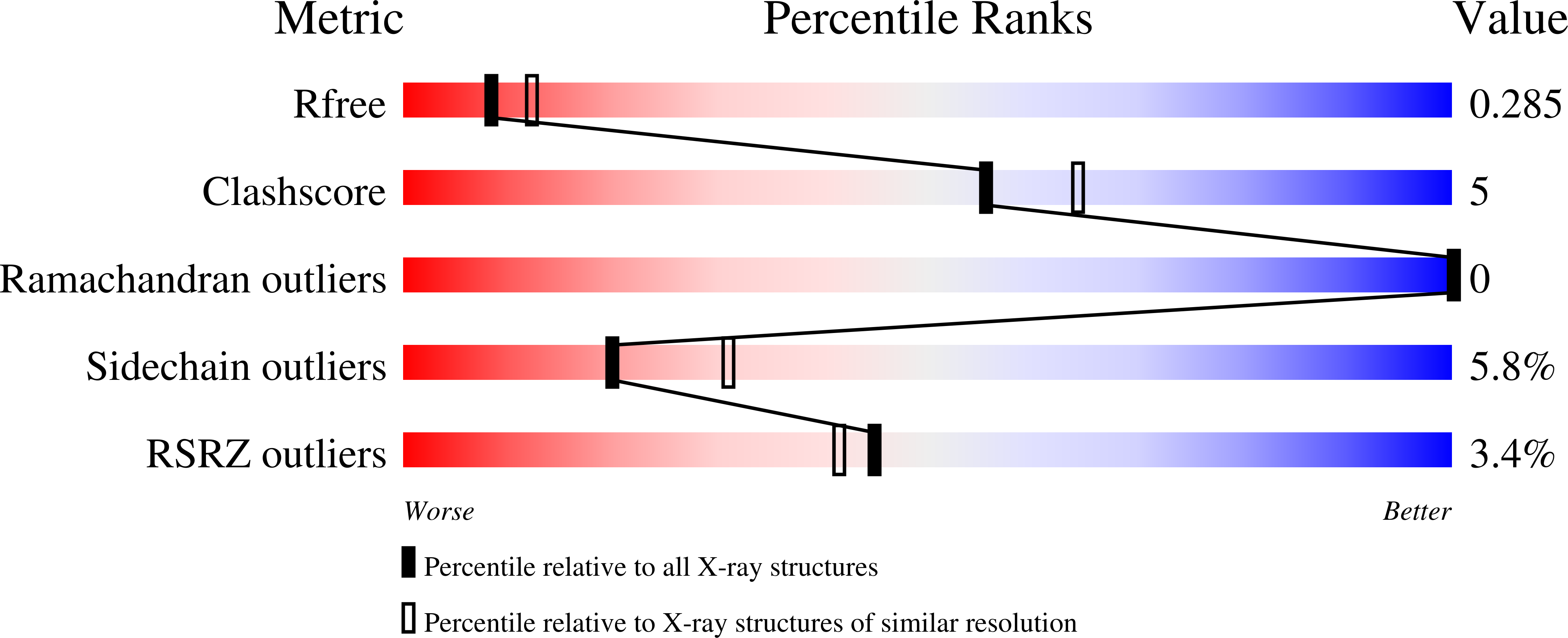
6XIH - PubMed Abstract:
Apoptosis Signal-Regulating Kinase-1 (ASK1) is a known member of the Mitogen-Activated Protein Kinase Kinase Kinase (MAP3K) family and upon stimulation will activate the p38- and JNK-pathways leading to cardiac apoptosis, fibrosis, and hypertrophy. Using Structure-Based Drug Design (SBDD) in parallel with deconstruction of a published compound, a novel series of ASK1 inhibitors was optimized, which incorporated a saturated heterocycle proximal to the hinge-binding motif. This yielded a unique chemical series with excellent selectivity across the broader kinome, and desirable drug-like properties. The lead compound (10) is highly soluble and permeable, and exhibits a cellular EC 50 = 24 nM and K d < 1 nM. Of the 350 kinases tested, 10 has an IC 50 ≤ 500 nM for only eight of them. This paper will describe the design hypotheses behind this series, key data points during the optimization phase, as well as a possible structural rationale for the kinome selectivity. Based on crystallographic data, the presence of an aliphatic cycle adjacent to the hinge-binder in the active site of the protein kinase showed up in <1% of the >5000 structures in the Protein Data Bank, potentially conferring the selectivity seen in this series.
Organizational Affiliation:
Medicinal Chemistry & In Vitro Pharmacology, Gastroenterology Drug Discovery Unit, Takeda Research in California, 9625 Towne Centre Drive, San Diego, CA 92121, United States. Electronic address: simone.bigi@takeda.com.