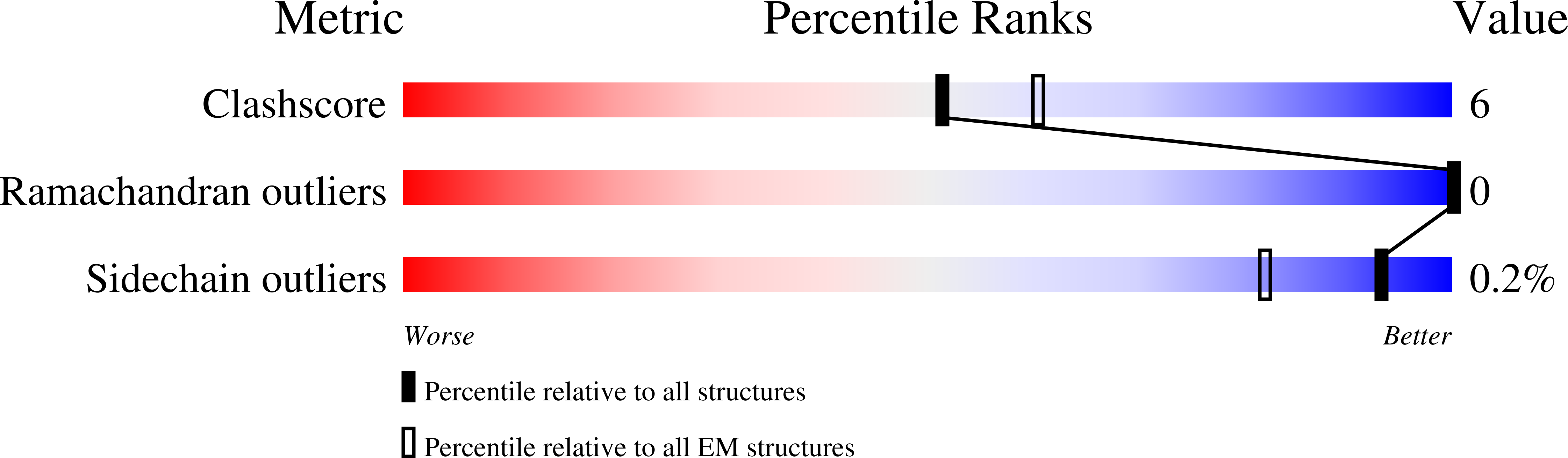Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Darricarrere, N., Qiu, Y., Kanekiyo, M., Creanga, A., Gillespie, R.A., Moin, S.M., Saleh, J., Sancho, J., Chou, T.H., Zhou, Y., Zhang, R., Dai, S., Moody, A., Saunders, K.O., Crank, M.C., Mascola, J.R., Graham, B.S., Wei, C.J., Nabel, G.J.(2021) Sci Transl Med 13
- PubMed: 33658355
- DOI: https://doi.org/10.1126/scitranslmed.abe5449
- Primary Citation of Related Structures:
6WZT, 6XGC - PubMed Abstract:
Seasonal influenza vaccines confer protection against specific viral strains but have restricted breadth that limits their protective efficacy. The H1 and H3 subtypes of influenza A virus cause most of the seasonal epidemics observed in humans and are the major drivers of influenza A virus-associated mortality. The consequences of pandemic spread of COVID-19 underscore the public health importance of prospective vaccine development. Here, we show that headless hemagglutinin (HA) stabilized-stem immunogens presented on ferritin nanoparticles elicit broadly neutralizing antibody (bnAb) responses to diverse H1 and H3 viruses in nonhuman primates (NHPs) when delivered with a squalene-based oil-in-water emulsion adjuvant, AF03. The neutralization potency and breadth of antibodies isolated from NHPs were comparable to human bnAbs and extended to mismatched heterosubtypic influenza viruses. Although NHPs lack the immunoglobulin germline VH1-69 residues associated with the most prevalent human stem-directed bnAbs, other gene families compensated to generate bnAbs. Isolation and structural analyses of vaccine-induced bnAbs revealed extensive interaction with the fusion peptide on the HA stem, which is essential for viral entry. Antibodies elicited by these headless HA stabilized-stem vaccines neutralized diverse H1 and H3 influenza viruses and shared a mode of recognition analogous to human bnAbs, suggesting that these vaccines have the potential to confer broadly protective immunity against diverse viruses responsible for seasonal and pandemic influenza infections in humans.
Organizational Affiliation:
Sanofi, 640 Memorial Drive, Cambridge, MA 02139, USA.