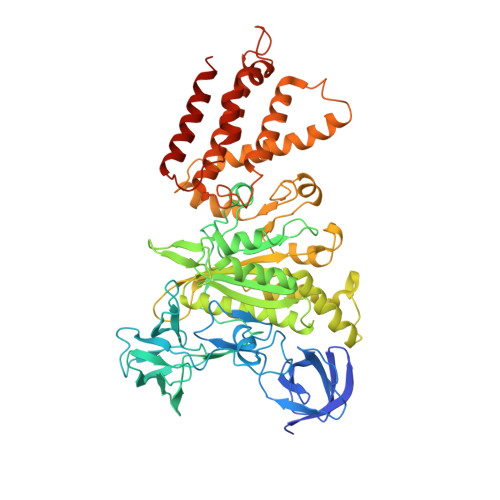
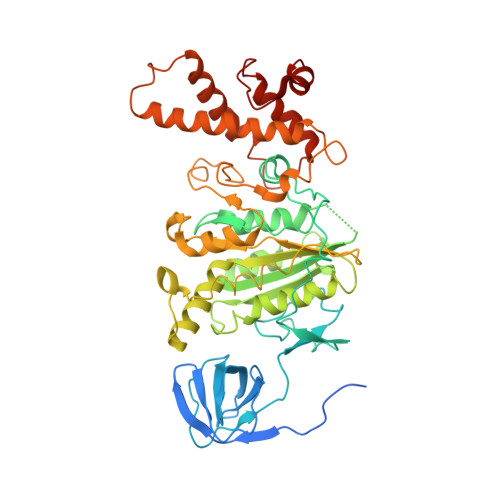
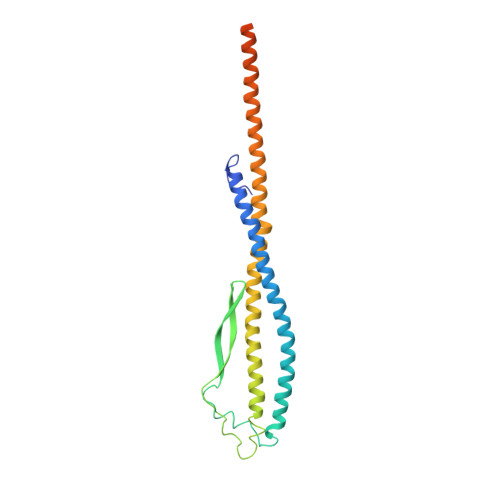
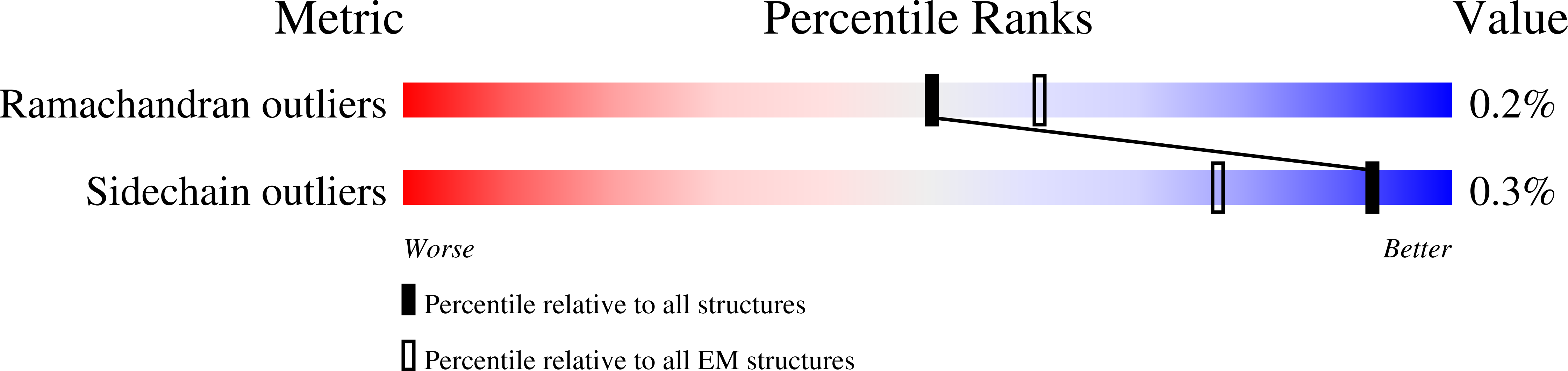Cryo-EM structures of intact V-ATPase from bovine brain.
Wang, R., Long, T., Hassan, A., Wang, J., Sun, Y., Xie, X.S., Li, X.(2020) Nat Commun 11: 3921-3921
- PubMed: 32764564
- DOI: https://doi.org/10.1038/s41467-020-17762-9
- Primary Citation of Related Structures:
6XBW, 6XBY - PubMed Abstract:
The vacuolar-type H + -ATPases (V-ATPase) hydrolyze ATP to pump protons across the plasma or intracellular membrane, secreting acids to the lumen or acidifying intracellular compartments. It has been implicated in tumor metastasis, renal tubular acidosis, and osteoporosis. Here, we report two cryo-EM structures of the intact V-ATPase from bovine brain with all the subunits including the subunit H, which is essential for ATPase activity. Two type-I transmembrane proteins, Ac45 and (pro)renin receptor, along with subunit c", constitute the core of the c-ring. Three different conformations of A/B heterodimers suggest a mechanism for ATP hydrolysis that triggers a rotation of subunits DF, inducing spinning of subunit d with respect to the entire c-ring. Moreover, many lipid molecules have been observed in the Vo domain to mediate the interactions between subunit c, c", (pro)renin receptor, and Ac45. These two structures reveal unique features of mammalian V-ATPase and suggest a mechanism of V1-Vo torque transmission.
Organizational Affiliation:
Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX, 75390, USA.