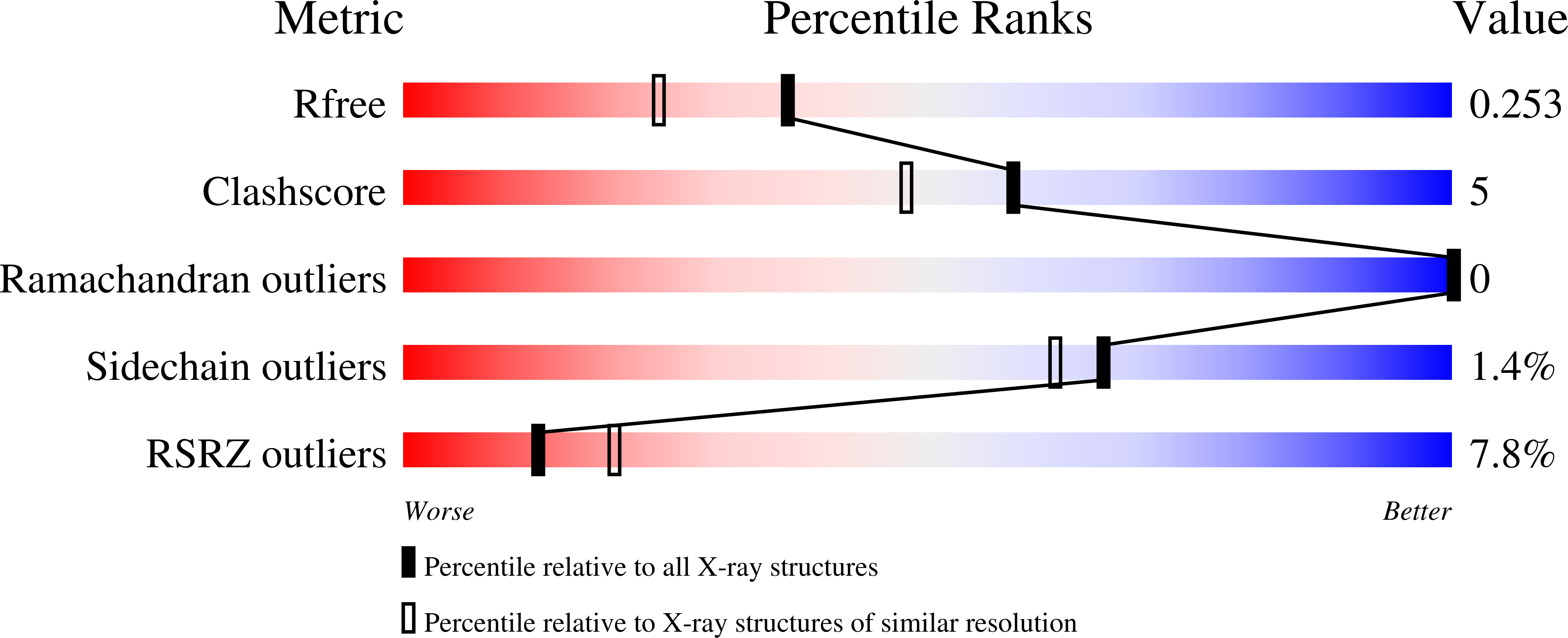
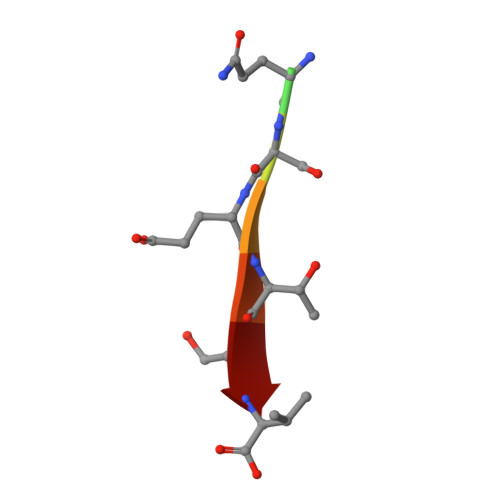
Structural basis of the human Scribble-Vangl2 association in health and disease.
How, J.Y., Stephens, R.K., Lim, K.Y.B., Humbert, P.O., Kvansakul, M.(2021) Biochem J 478: 1321-1332
- PubMed: 33684218
- DOI: https://doi.org/10.1042/BCJ20200816
- Primary Citation of Related Structures:
6XA6, 6XA7, 6XA8, 7JO7 - PubMed Abstract:
Scribble is a critical cell polarity regulator that has been shown to work as either an oncogene or tumor suppressor in a context dependent manner, and also impacts cell migration, tissue architecture and immunity. Mutations in Scribble lead to neural tube defects in mice and humans, which has been attributed to a loss of interaction with the planar cell polarity regulator Vangl2. We show that the Scribble PDZ domains 1, 2 and 3 are able to interact with the C-terminal PDZ binding motif of Vangl2 and have now determined crystal structures of these Scribble PDZ domains bound to the Vangl2 peptide. Mapping of mammalian neural tube defect mutations reveal that mutations located distal to the canonical PDZ domain ligand binding groove can not only ablate binding to Vangl2 but also disrupt binding to multiple other signaling regulators. Our findings suggest that PDZ-associated neural tube defect mutations in Scribble may not simply act in a Vangl2 dependent manner but as broad-spectrum loss of function mutants by disrupting the global Scribble-mediated interaction network.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria 3086, Australia.