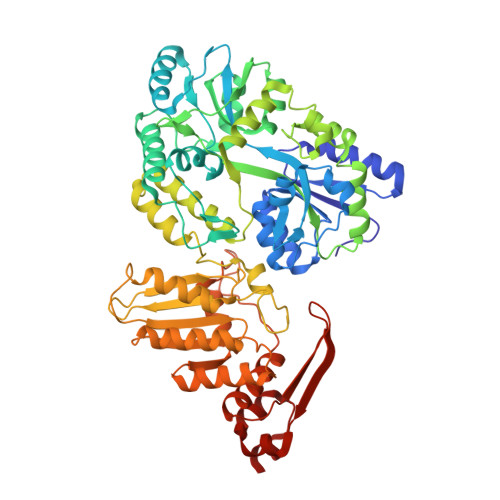
The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.
Wolfe, A.D., Li, S., Goedderz, C., Chen, X.S.(2020) NAR Cancer 2: zcaa027-zcaa027
- PubMed: 33094286
- DOI: https://doi.org/10.1093/narcan/zcaa027
- Primary Citation of Related Structures:
6X91 - PubMed Abstract:
APOBEC1 (APO1), a member of AID/APOBEC nucleic acid cytosine deaminase family, can edit apolipoprotein B mRNA to regulate cholesterol metabolism. This APO1 RNA editing activity requires a cellular cofactor to achieve tight regulation. However, no cofactors are required for deamination on DNA by APO1 and other AID/APOBEC members, and aberrant deamination on genomic DNA by AID/APOBEC deaminases has been linked to cancer. Here, we present the crystal structure of APO1, which reveals a typical APOBEC deaminase core structure, plus a unique well-folded C-terminal domain that is highly hydrophobic. This APO1 C-terminal hydrophobic domain (A1HD) interacts to form a stable dimer mainly through hydrophobic interactions within the dimer interface to create a four-stranded β-sheet positively charged surface. Structure-guided mutagenesis within this and other regions of APO1 clarified the importance of the A1HD in directing RNA and cofactor interactions, providing insights into the structural basis of selectivity on DNA or RNA substrates.
Organizational Affiliation:
Genetics, Molecular and Cellular Biology, Keck School of Medicine, University of Southern California, Los Angeles, CA 90089, USA.