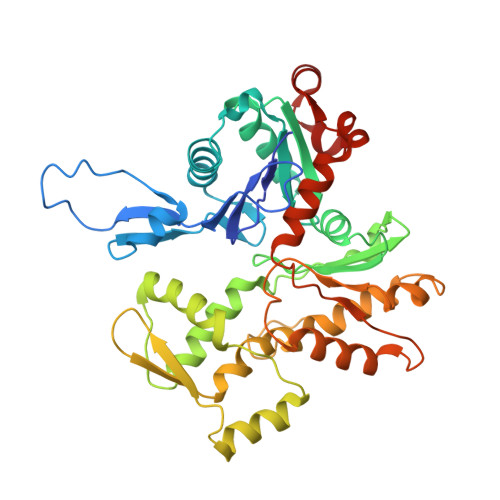
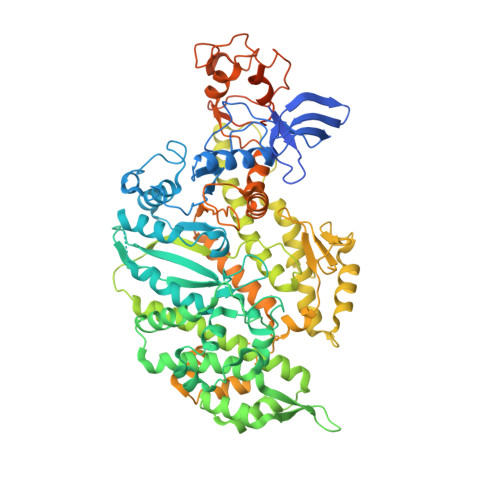
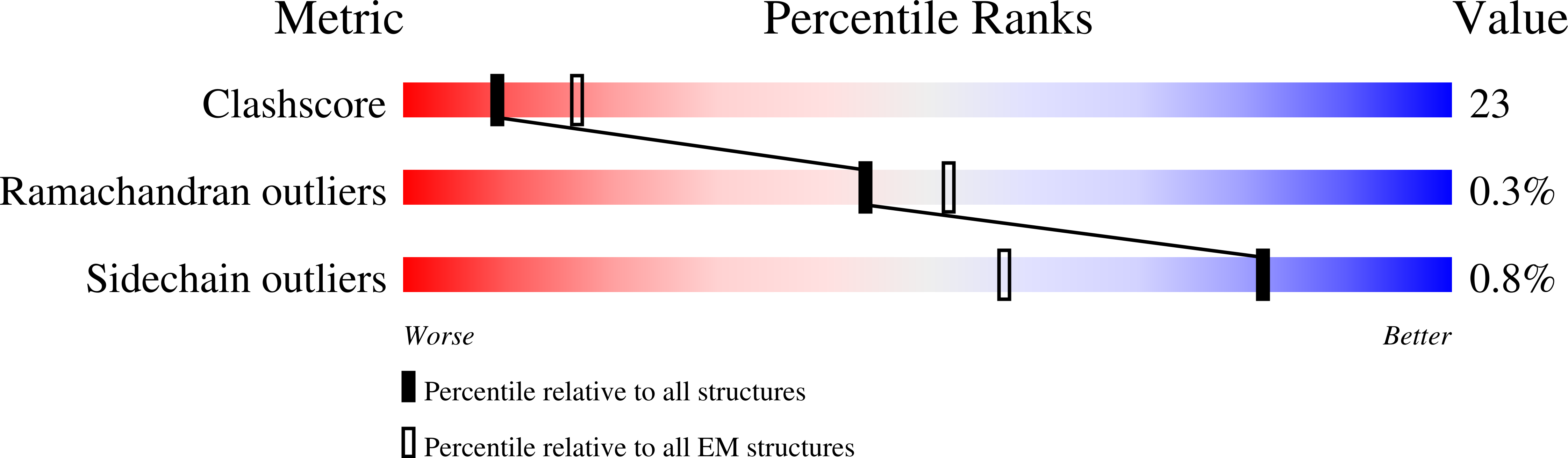Cryo-EM and Molecular Docking Shows Myosin Loop 4 Contacts Actin and Tropomyosin on Thin Filaments.
Doran, M.H., Pavadai, E., Rynkiewicz, M.J., Walklate, J., Bullitt, E., Moore, J.R., Regnier, M., Geeves, M.A., Lehman, W.(2020) Biophys J 119: 821-830
- PubMed: 32730789
- DOI: https://doi.org/10.1016/j.bpj.2020.07.006
- Primary Citation of Related Structures:
6X5Z - PubMed Abstract:
The motor protein myosin drives muscle and nonmuscle motility by binding to and moving along actin of thin filaments. Myosin binding to actin also modulates interactions of the regulatory protein, tropomyosin, on thin filaments, and conversely tropomyosin affects myosin binding to actin. Insight into this reciprocity will facilitate a molecular level elucidation of tropomyosin regulation of myosin interaction with actin in muscle contraction, and in turn, promote better understanding of nonmuscle cell motility. Indeed, experimental approaches such as fiber diffraction, cryoelectron microscopy, and three-dimensional reconstruction have long been used to define regulatory interaction of tropomyosin and myosin on actin at a structural level. However, their limited resolution has not proven sufficient to determine tropomyosin and myosin contacts at an atomic-level and thus to fully substantiate possible functional contributions. To overcome this deficiency, we have followed a hybrid approach by performing new cryogenic electron microscopy reconstruction of myosin-S1-decorated F-actin-tropomyosin together with atomic scale protein-protein docking of tropomyosin to the EM models. Here, cryo-EM data were derived from filaments reconstituted with α1-actin, cardiac αα-tropomyosin, and masseter muscle β-myosin complexes; masseter myosin, which shares sequence identity with β-cardiac myosin-heavy chain, was used because of its stability in vitro. The data were used to build an atomic model of the tropomyosin cable that fits onto the actin filament between the tip of the myosin head and a cleft on the innermost edge of actin subunits. The docking and atomic scale fitting showed multiple discrete interactions of myosin loop 4 and acidic residues on successive 39-42 residue-long tropomyosin pseudorepeats. The contacts between S1 and tropomyosin on actin appear to compete with and displace ones normally found between actin and tropomyosin on myosin-free thin filaments in relaxed muscle, thus restructuring the filament during myosin-induced activation.
Organizational Affiliation:
Department of Physiology & Biophysics, Boston University School of Medicine, Boston, Massachusetts.