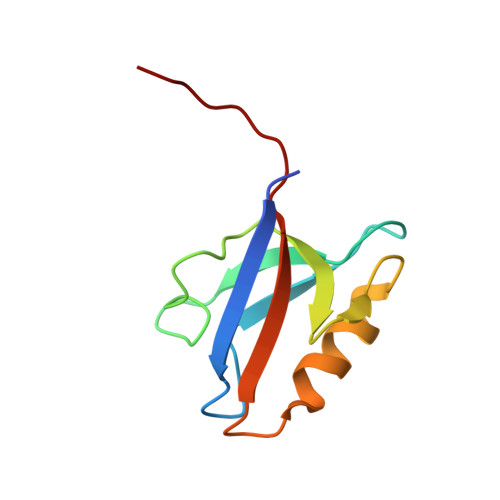
Structural characterization and computational analysis of PDZ domains in Monosiga brevicollis.
Gao, M., Mackley, I.G.P., Mesbahi-Vasey, S., Bamonte, H.A., Struyvenberg, S.A., Landolt, L., Pederson, N.J., Williams, L.I., Bahl, C.D., Brooks 3rd, L., Amacher, J.F.(2020) Protein Sci 29: 2226-2244
- PubMed: 32914530
- DOI: https://doi.org/10.1002/pro.3947
- Primary Citation of Related Structures:
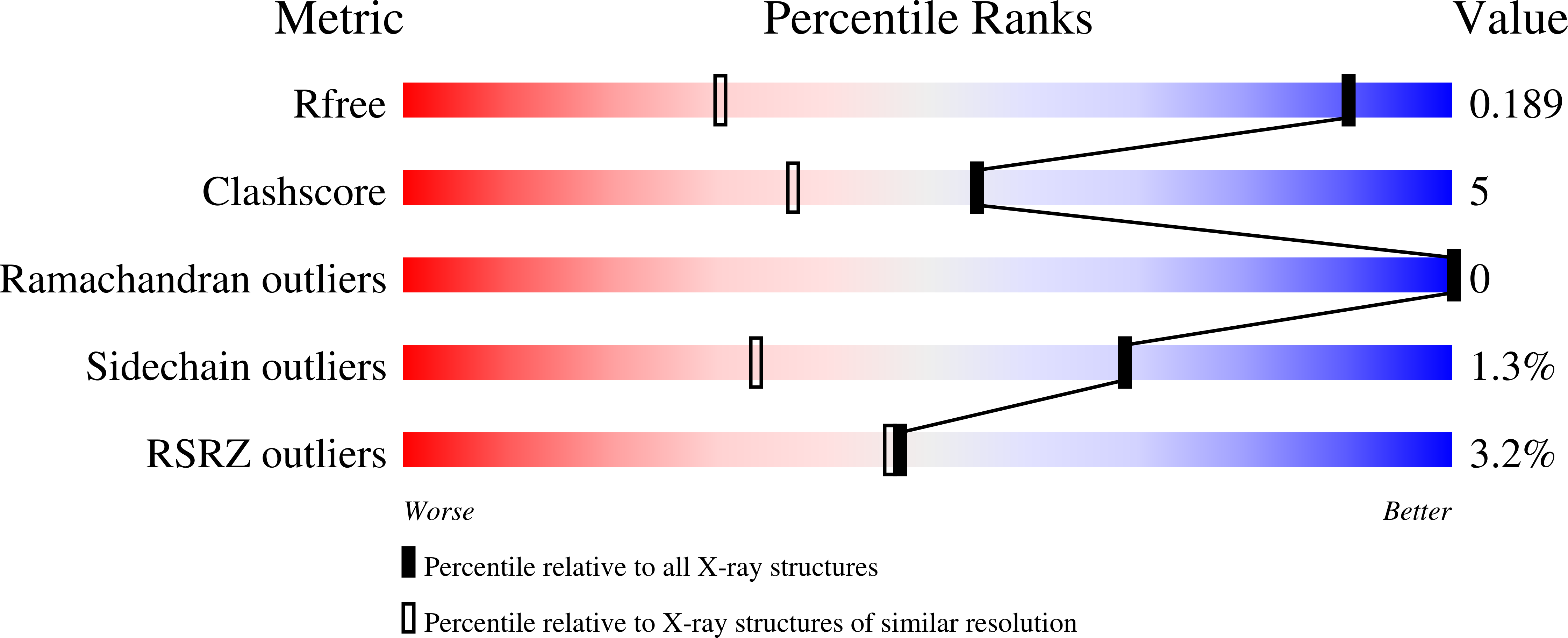
6X1N, 6X1P, 6X1R, 6X1X, 6X20, 6X22, 6X23 - PubMed Abstract:
Identification of the molecular networks that facilitated the evolution of multicellular animals from their unicellular ancestors is a fundamental problem in evolutionary cellular biology. Choanoflagellates are recognized as the closest extant nonmetazoan ancestors to animals. These unicellular eukaryotes can adopt a multicellular-like "rosette" state. Therefore, they are compelling models for the study of early multicellularity. Comparative studies revealed that a number of putative human orthologs are present in choanoflagellate genomes, suggesting that a subset of these genes were necessary for the emergence of multicellularity. However, previous work is largely based on sequence alignments alone, which does not confirm structural nor functional similarity. Here, we focus on the PDZ domain, a peptide-binding domain which plays critical roles in myriad cellular signaling networks and which underwent a gene family expansion in metazoan lineages. Using a customized sequence similarity search algorithm, we identified 178 PDZ domains in the Monosiga brevicollis proteome. This includes 11 previously unidentified sequences, which we analyzed using Rosetta and homology modeling. To assess conservation of protein structure, we solved high-resolution crystal structures of representative M. brevicollis PDZ domains that are homologous to human Dlg1 PDZ2, Dlg1 PDZ3, GIPC, and SHANK1 PDZ domains. To assess functional conservation, we calculated binding affinities for mbGIPC, mbSHANK1, mbSNX27, and mbDLG-3 PDZ domains from M. brevicollis. Overall, we find that peptide selectivity is generally conserved between these two disparate organisms, with one possible exception, mbDLG-3. Overall, our results provide novel insight into signaling pathways in a choanoflagellate model of primitive multicellularity.
Organizational Affiliation:
Department of Chemistry, Western Washington University, Bellingham, Washington, USA.