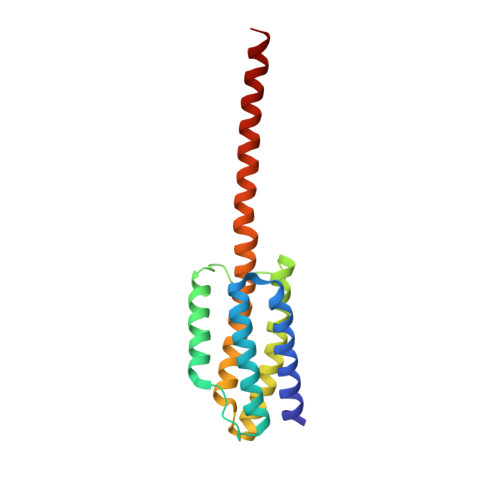
Geometric Lessons and Design Strategies for Nanoscale Protein Cages.
Laniado, J., Cannon, K.A., Miller, J.E., Sawaya, M.R., McNamara, D.E., Yeates, T.O.(2021) ACS Nano 15: 4277-4286
- PubMed: 33683103
- DOI: https://doi.org/10.1021/acsnano.0c07167
- Primary Citation of Related Structures:
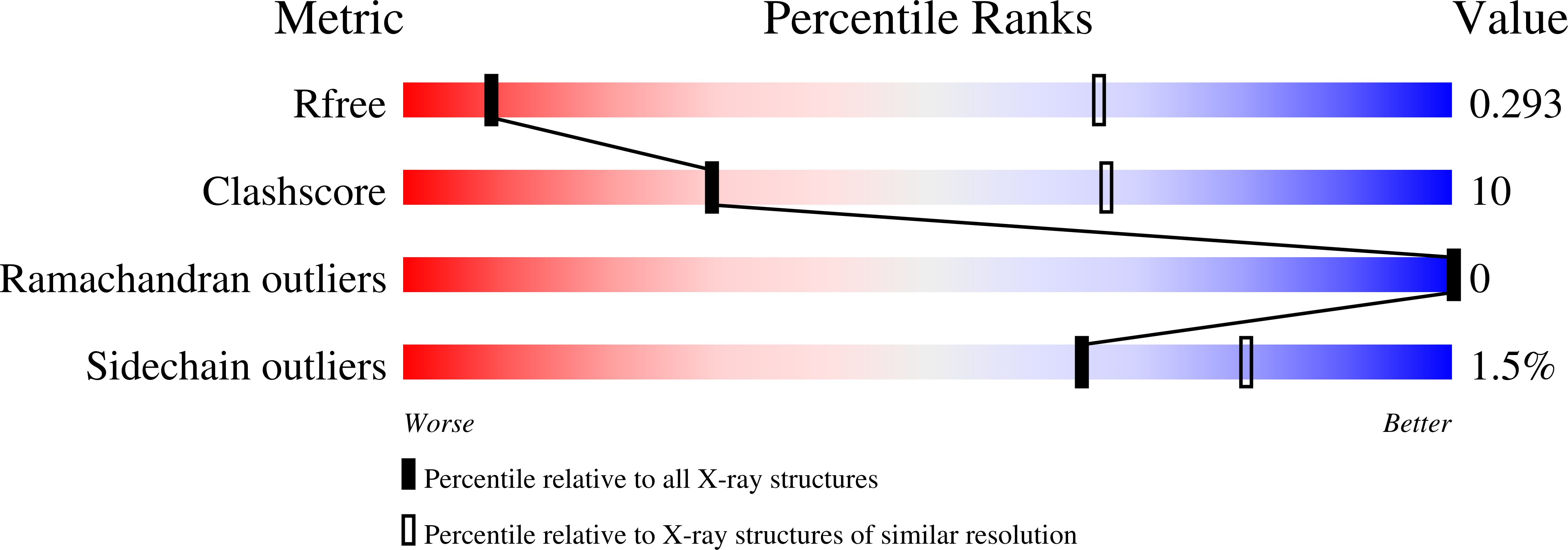
6X1I - PubMed Abstract:
Protein molecules bring a rich functionality to the field of designed nanoscale architectures. High-symmetry protein cages are rapidly finding diverse applications in biomedicine, nanotechnology, and imaging, but methods for their reliable and predictable construction remain challenging. In this study we introduce an approach for designing protein assemblies that combines ideas and favorable elements adapted from recent work. Cubically symmetric cages can be created by combining two simpler symmetries, following recently established principles. Here, two different oligomeric protein components are brought together in a geometrically specific arrangement by their separate genetic fusion to individual components of a heterodimeric coiled-coil polypeptide motif of known structure. Fusions between components are made by continuous α-helices to limit flexibility. After a computational design, we tested 10 different protein cage constructions experimentally, two of which formed larger assemblies. One produced the intended octahedral cage, ∼26 nm in diameter, while the other appeared to produce the intended tetrahedral cage as a minor component, crystallizing instead in an alternate form representing a collapsed structure of lower stoichiometry and symmetry. Geometric distinctions between the two characterized designs help explain the different degrees of success, leading to clearer principles and improved prospects for the routine creation of nanoscale protein architectures using diverse methods.
Organizational Affiliation:
Molecular Biology Institute, UCLA, Los Angeles, California 90095, United States.