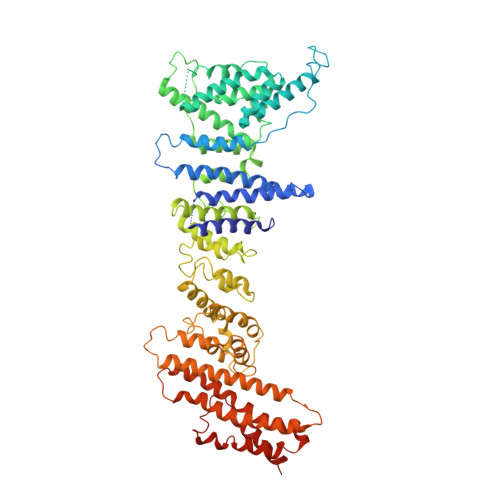
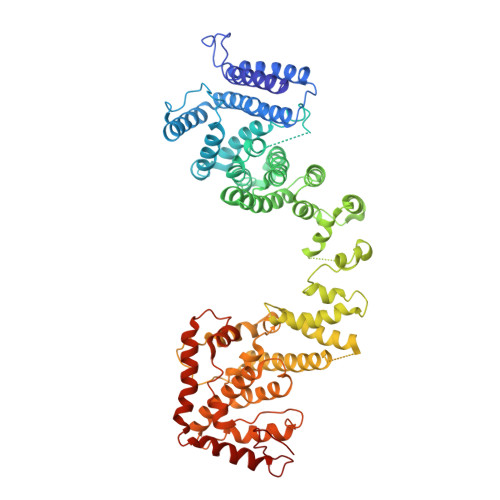
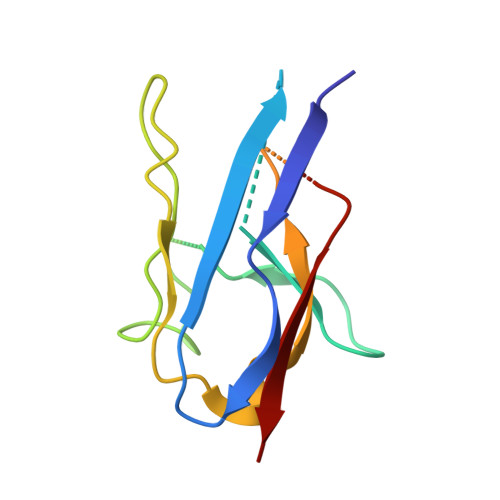
Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif.
Nordeen, S.A., Turman, D.L., Schwartz, T.U.(2020) Nat Commun 11: 6060-6060
- PubMed: 33247142
- DOI: https://doi.org/10.1038/s41467-020-19885-5
- Primary Citation of Related Structures:
6X02, 6X03, 6X04, 6X05 - PubMed Abstract:
The hallmark of the eukaryotic cell is the complex endomembrane system that compartmentalizes cellular functions. Transport into and out of the nucleus occurs through the nuclear pore complex (NPC). The heptameric Nup84 or Y complex is an essential scaffolding component of the NPC. Here we report two nanobody-bound structures: the full-length Nup84-Nup133 C-terminal domain complex and the Nup133 N-terminal domain, both from S. cerevisiae. Together with previously published structures, this work enables the structural description of the entire 575 kDa Y complex from one species. The structure of Nup84-Nup133 CTD details the high flexibility of this dimeric unit of the Y complex. Further, the Nup133 NTD contains a structurally conserved amphipathic lipid packing sensor motif, confirmed by liposome interaction studies. The presented structures reveal important details about the function of the Y complex that affect our understanding of NPC structure and assembly.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.