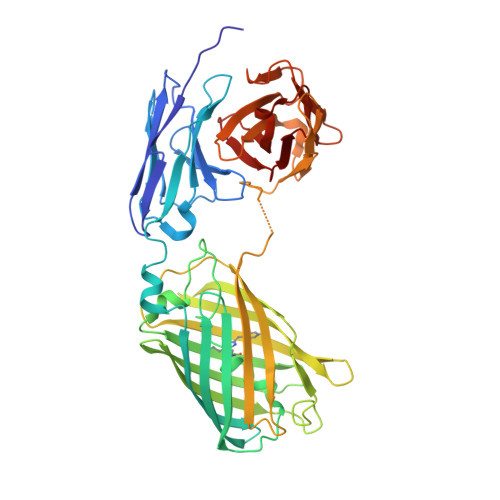
Construction, characterization and crystal structure of a fluorescent single-chain Fv chimera.
Velappan, N., Close, D., Hung, L.W., Naranjo, L., Hemez, C., DeVore, N., McCullough, D.K., Lillo, A.M., Waldo, G.S., Bradbury, A.R.M.(2021) Protein Eng Des Sel 34
- PubMed: 33586761
- DOI: https://doi.org/10.1093/protein/gzaa029
- Primary Citation of Related Structures:
6WZN - PubMed Abstract:
In vitro display technologies based on phage and yeast have a successful history of selecting single-chain variable fragment (scFv) antibodies against various targets. However, single-chain antibodies are often unstable and poorly expressed in Escherichia coli. Here, we explore the feasibility of converting scFv antibodies to an intrinsically fluorescent format by inserting the monomeric, stable fluorescent protein named thermal green, between the light- and heavy-chain variable regions. Our results show that the scTGP format maintains the affinity and specificity of the antibodies, improves expression levels, allows one-step fluorescent assay for detection of binding and is a suitable reagent for epitope binning. We also report the crystal structure of an scTGP construct that recognizes phosphorylated tyrosine on FcεR1 receptor of the allergy pathway.
Organizational Affiliation:
Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87545, USA.