Mutational fitness landscapes reveal genetic and structural improvement pathways for a vaccine-elicited HIV-1 broadly neutralizing antibody.
Madan, B., Zhang, B., Xu, K., Chao, C.W., O'Dell, S., Wolfe, J.R., Chuang, G.Y., Fahad, A.S., Geng, H., Kong, R., Louder, M.K., Nguyen, T.D., Rawi, R., Schon, A., Sheng, Z., Nimrania, R., Wang, Y., Zhou, T., Lin, B.C., Doria-Rose, N.A., Shapiro, L., Kwong, P.D., DeKosky, B.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33649208
- DOI: https://doi.org/10.1073/pnas.2011653118
- Primary Citation of Related Structures:
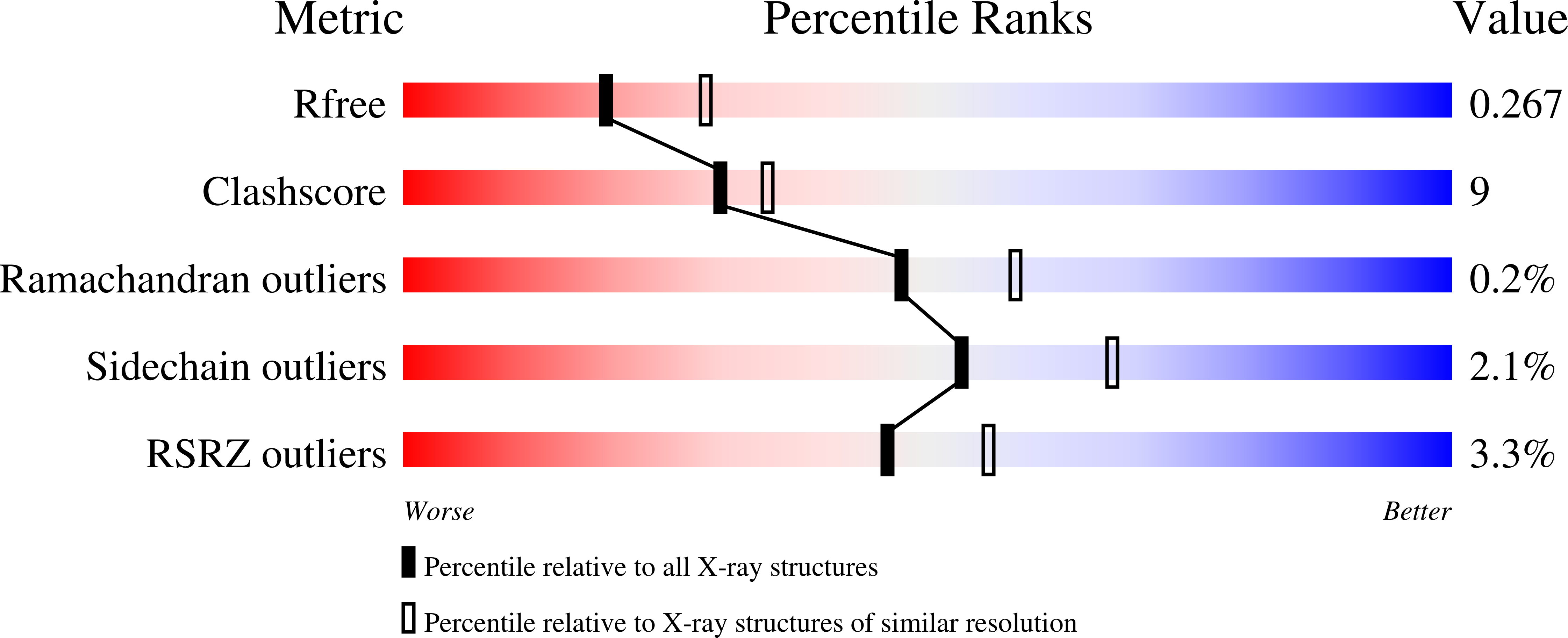
6WWC, 6WX2 - PubMed Abstract:
Vaccine-based elicitation of broadly neutralizing antibodies holds great promise for preventing HIV-1 transmission. However, the key biophysical markers of improved antibody recognition remain uncertain in the diverse landscape of potential antibody mutation pathways, and a more complete understanding of anti-HIV-1 fusion peptide (FP) antibody development will accelerate rational vaccine designs. Here we survey the mutational landscape of the vaccine-elicited anti-FP antibody, vFP16.02, to determine the genetic, structural, and functional features associated with antibody improvement or fitness. Using site-saturation mutagenesis and yeast display functional screening, we found that 1% of possible single mutations improved HIV-1 envelope trimer (Env) affinity, but generally comprised rare somatic hypermutations that may not arise frequently in vivo. We observed that many single mutations in the vFP16.02 Fab could enhance affinity >1,000-fold against soluble FP, although affinity improvements against the HIV-1 trimer were more measured and rare. The most potent variants enhanced affinity to both soluble FP and Env, had mutations concentrated in antibody framework regions, and achieved up to 37% neutralization breadth compared to 28% neutralization of the template antibody. Altered heavy- and light-chain interface angles and conformational dynamics, as well as reduced Fab thermal stability, were associated with improved HIV-1 neutralization breadth and potency. We also observed parallel sets of mutations that enhanced viral neutralization through similar structural mechanisms. These data provide a quantitative understanding of the mutational landscape for vaccine-elicited FP-directed broadly neutralizing antibody and demonstrate that numerous antigen-distal framework mutations can improve antibody function by enhancing affinity simultaneously toward HIV-1 Env and FP.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, The University of Kansas, Lawrence, KS 66045.