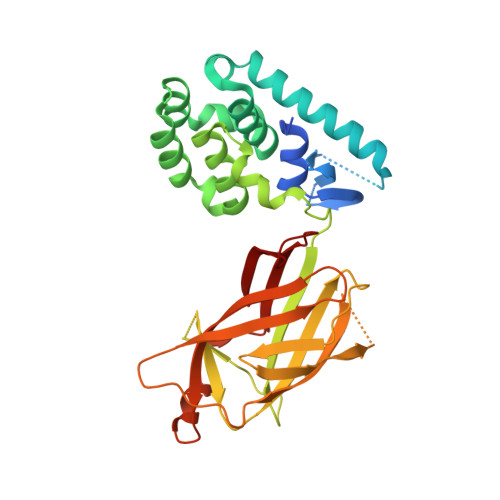
Cannabinoid receptor interacting protein 1a interacts with myristoylated G alpha i N terminus via a unique gapped beta-barrel structure.
Booth, W.T., Clodfelter, J.E., Leone-Kabler, S., Hughes, E.K., Eldeeb, K., Howlett, A.C., Lowther, W.T.(2021) J Biol Chem 297: 101099-101099
- PubMed: 34418434
- DOI: https://doi.org/10.1016/j.jbc.2021.101099
- Primary Citation of Related Structures:
6WSK - PubMed Abstract:
Cannabinoid receptor interacting protein 1a (CRIP1a) modulates CB 1 cannabinoid receptor G-protein coupling in part by altering the selectivity for Gα i subtype activation, but the molecular basis for this function of CRIP1a is not known. We report herein the first structure of CRIP1a at a resolution of 1.55 Å. CRIP1a exhibits a 10-stranded and antiparallel β-barrel with an interior comprised of conserved hydrophobic residues and loops at the bottom and a short helical cap at the top to exclude solvent. The β-barrel has a gap between strands β8 and β10, which deviates from β-sandwich fatty acid-binding proteins that carry endocannabinoid compounds and the Rho-guanine nucleotide dissociation inhibitor predicted by computational threading algorithms. The structural homology search program DALI identified CRIP1a as homologous to a family of lipidated-protein carriers that includes phosphodiesterase 6 delta subunit and Unc119. Comparison with these proteins suggests that CRIP1a may carry two possible types of cargo: either (i) like phosphodiesterase 6 delta subunit, cargo with a farnesyl moiety that enters from the top of the β-barrel to occupy the hydrophobic interior or (ii) like Unc119, cargo with a palmitoyl or a myristoyl moiety that enters from the side where the missing β-strand creates an opening to the hydrophobic pocket. Fluorescence polarization analysis demonstrated CRIP1a binding of an N-terminally myristoylated 9-mer peptide mimicking the Gα i N terminus. However, CRIP1a could not bind the nonmyristolyated Gα i peptide or cargo of homologs. Thus, binding of CRIP1a to Gαi proteins represents a novel mechanism to regulate cell signaling initiated by the CB 1 receptor.
Organizational Affiliation:
Department of Biochemistry and Center for Structural Biology, Wake Forest School of Medicine, Winston-Salem, North Carolina, USA.