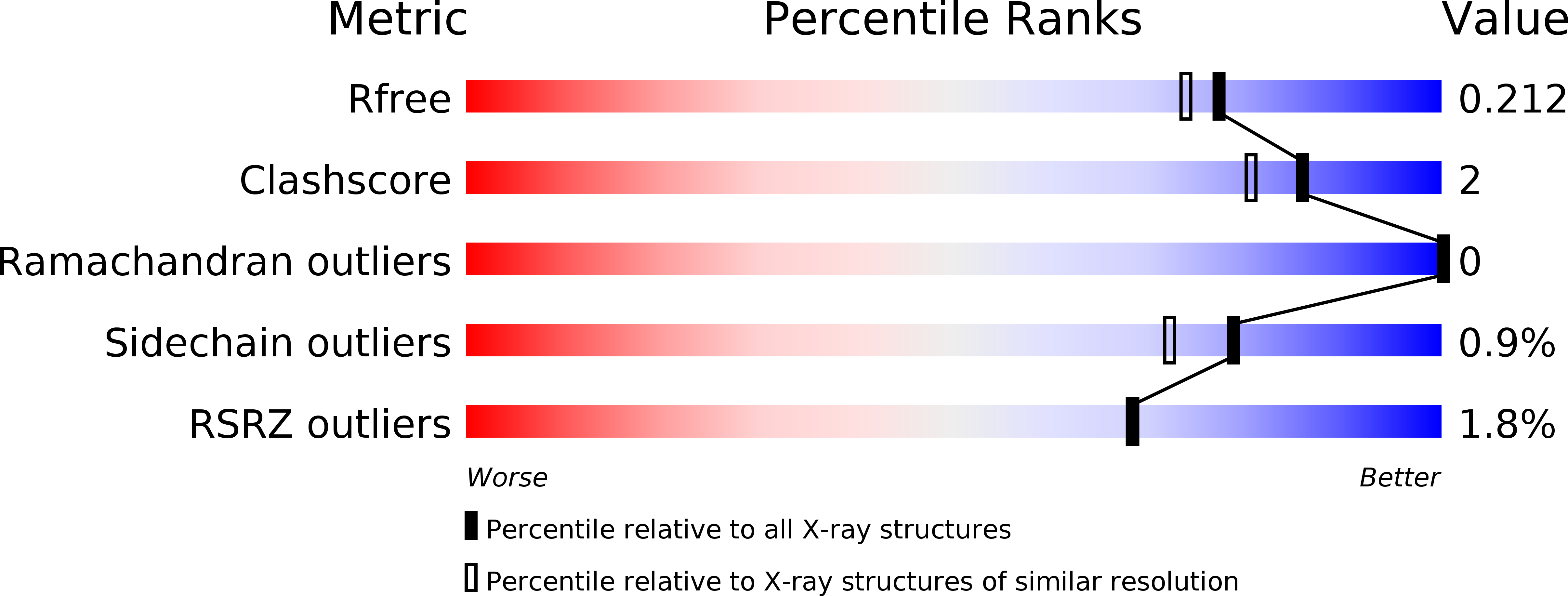
The development of a structurally distinct series of BACE1 inhibitors via the (Z)-fluoro-olefin amide bioisosteric replacement.
Frohn, M., Liu, L., Siegmund, A.C., Qian, W., Amegadzie, A., Chen, N., Tan, H., Hickman, D., Wood, S., Wen, P.H., Bartberger, M.D., Whittington, D.A., Allen, J.R., Bourbeau, M.P.(2020) Bioorg Med Chem Lett 30: 127240-127240
- PubMed: 32527542
- DOI: https://doi.org/10.1016/j.bmcl.2020.127240
- Primary Citation of Related Structures:
6WNY - PubMed Abstract:
The (Z)-fluoro-olefin amide bioisosteric replacement is an effective tool for addressing various shortcomings of the parent amide. In an effort to fine tune ADME properties of BACE1 preclinical candidate AM-6494, a series of structurally distinct (Z)-fluoro-olefin containing analogs was developed that culminated in compound 15. Herein, we detail design considerations, synthetic challenges, structure activity relationship (SAR) studies, and in vivo properties of an advanced compound in this novel series of BACE1 inhibitors.
Organizational Affiliation:
Medicinal Chemistry, Amgen Discovery Research, Amgen Inc., Thousand Oaks, CA 91320, United States. Electronic address: mfrohn@amgen.com.