In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Shan, L., Dyk, N.V., Haskins, N., Cook, K.M., Rosenthal, K.L., Mazor, R., Dragulin-Otto, S., Jiang, Y., Wu, H., Dall'Acqua, W.F., Borrok, M.J., Damschroder, M.M., Oganesyan, V.(2021) Commun Biol 4: 1048-1048
- PubMed: 34497355
- DOI: https://doi.org/10.1038/s42003-021-02565-5
- Primary Citation of Related Structures:
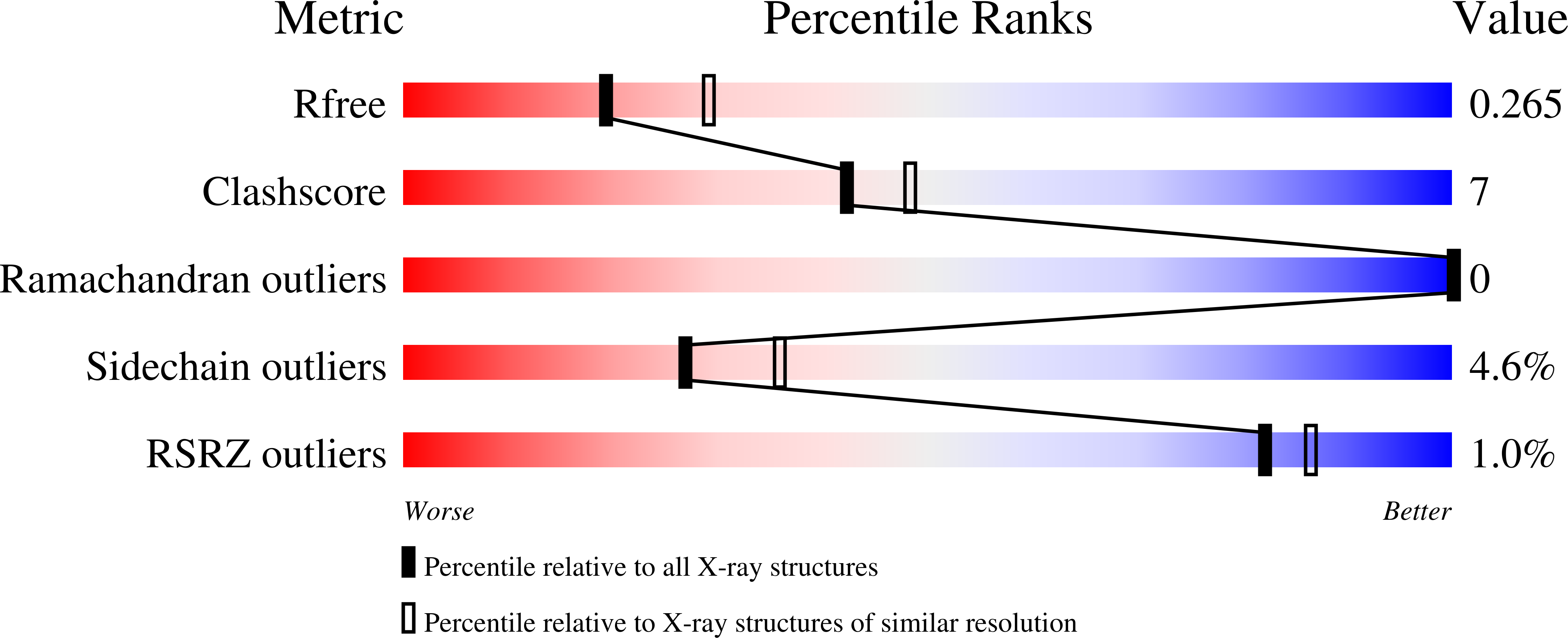
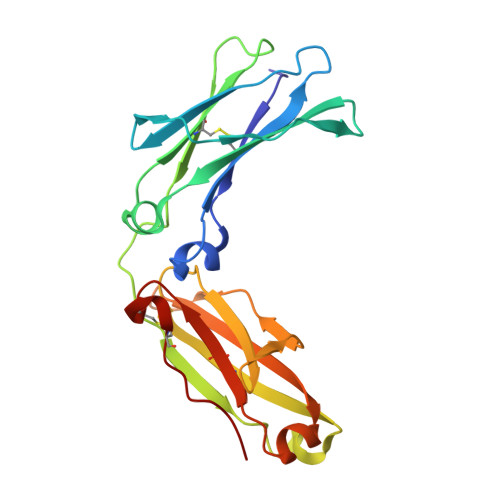
6WIB, 6WMH, 6WNA, 6WOL - PubMed Abstract:
In a biologic therapeutic landscape that requires versatility in targeting specificity, valency and half-life modulation, the monomeric Fc fusion platform holds exciting potential for the creation of a class of monovalent protein therapeutics that includes fusion proteins and bispecific targeting molecules. Here we report a structure-guided approach to engineer monomeric Fc molecules to adapt multiple versions of half-life extension modifications. Co-crystal structures of these monomeric Fc variants with Fc neonatal receptor (FcRn) shed light into the binding interactions that could serve as a guide for engineering the half-life of antibody Fc fragments. These engineered monomeric Fc molecules also enabled the generation of a novel monovalent bispecific molecular design, which translated the FcRn binding enhancement to improvement of in vivo serum half-life.
Organizational Affiliation:
Antibody Discovery and Protein Engineering, R&D, AstraZeneca, Gaithersburg, MD, USA. shan@dnli.com.