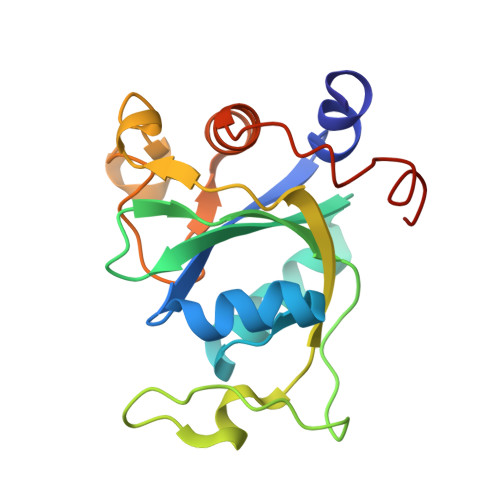
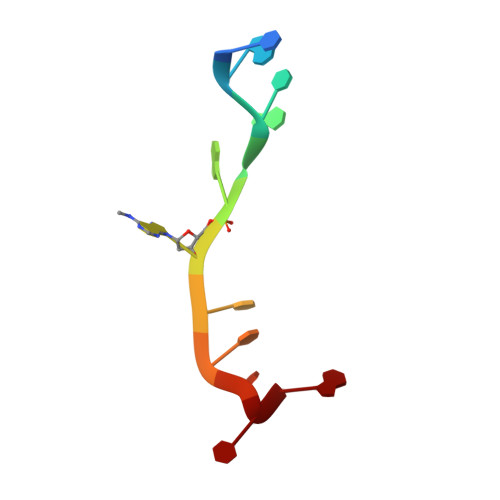
Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Woodcock, C.B., Horton, J.R., Zhou, J., Bedford, M.T., Blumenthal, R.M., Zhang, X., Cheng, X.(2020) Nucleic Acids Res 48: 10329-10341
- PubMed: 32663306
- DOI: https://doi.org/10.1093/nar/gkaa604
- Primary Citation of Related Structures:
6WE8, 6WE9, 6WEA - PubMed Abstract:
The recently characterized mammalian writer (methyltransferase) and eraser (demethylase) of the DNA N6-methyladenine (N6mA) methyl mark act on single-stranded (ss) and transiently-unpaired DNA. As YTH domain-containing proteins bind N6mA-containing RNA in mammalian cells, we investigated whether mammalian YTH domains are also methyl mark readers of N6mA DNA. Here, we show that the YTH domain of YTHDC1 (known to localize in the nucleus) binds ssDNA containing N6mA, with a 10 nM dissociation constant. This binding is stronger by a factor of 5 than in an RNA context, tested under the same conditions. However, the YTH domains of YTHDF2 and YTHDF1 (predominantly cytoplasmic) exhibited the opposite effect with ∼1.5-2× stronger binding to ssRNA containing N6mA than to the corresponding DNA. We determined two structures of the YTH domain of YTHDC1 in complex with N6mA-containing ssDNA, which illustrated that YTHDC1 binds the methylated adenine in a single-stranded region flanked by duplexed DNA. We discuss the hypothesis that the writer-reader-eraser of N6mA-containining ssDNA is associated with maintaining genome stability. Structural comparison of YTH and SRA domains (the latter a DNA 5-methylcytosine reader) revealed them to be diverse members of a larger family of DNA/RNA modification readers, apparently having originated from bacterial modification-dependent restriction enzymes.
Organizational Affiliation:
Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.