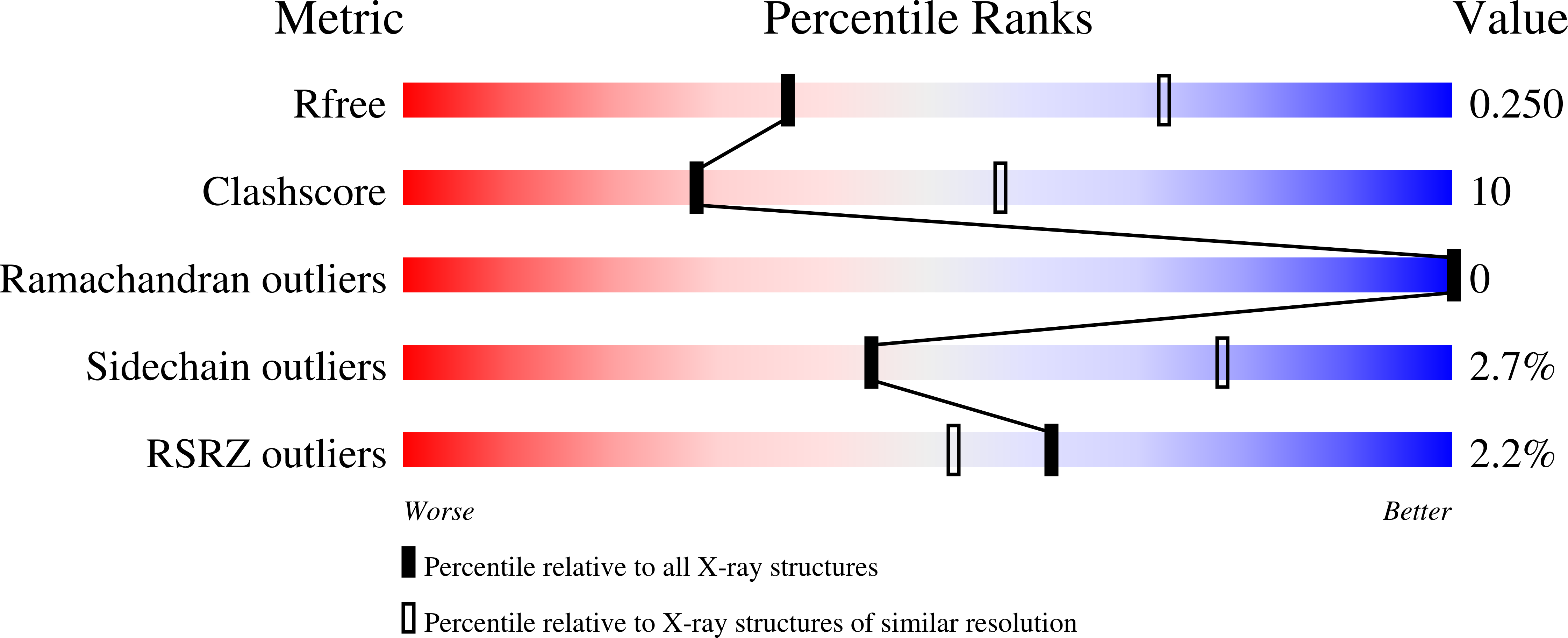
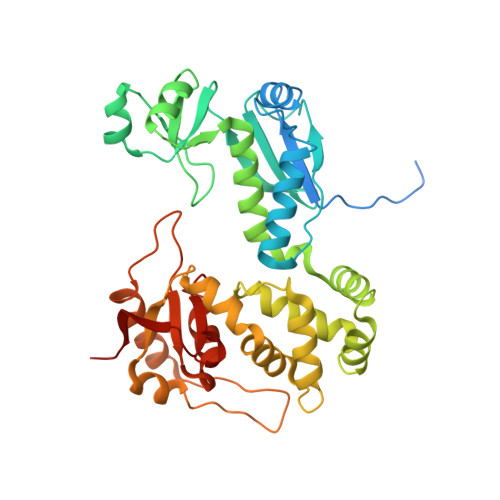
"The structure of the Type III secretion system export gate with CdsO, an ATPase lever arm".
Jensen, J.L., Yamini, S., Rietsch, A., Spiller, B.W.(2020) PLoS Pathog 16: e1008923-e1008923
- PubMed: 33048983
- DOI: https://doi.org/10.1371/journal.ppat.1008923
- Primary Citation of Related Structures:
6WA6, 6WA9 - PubMed Abstract:
Type III protein secretion systems (T3SS) deliver effector proteins from the Gram-negative bacterial cytoplasm into a eukaryotic host cell through a syringe-like, multi-protein nanomachine. Cytosolic components of T3SS include a portion of the export apparatus, which traverses the inner membrane and features the opening of the secretion channel, and the sorting complex for substrate recognition and for providing the energetics required for protein secretion. Two components critical for efficient effector export are the export gate protein and the ATPase, which are proposed to be linked by the central stalk protein of the ATPase. We present the structure of the soluble export gate homo-nonamer, CdsV, in complex with the central stalk protein, CdsO, of its cognate ATPase, both derived from Chlamydia pneumoniae. This structure defines the interface between these essential T3S proteins and reveals that CdsO engages the periphery of the export gate that may allow the ATPase to catalyze an opening between export gate subunits to allow cargo to enter the export apparatus. We also demonstrate through structure-based mutagenesis of the homologous export gate in Pseudomonas aeruginosa that mutation of this interface disrupts effector secretion. These results provide novel insights into the molecular mechanisms governing active substrate recognition and translocation through a T3SS.
Organizational Affiliation:
Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN, United States of America.