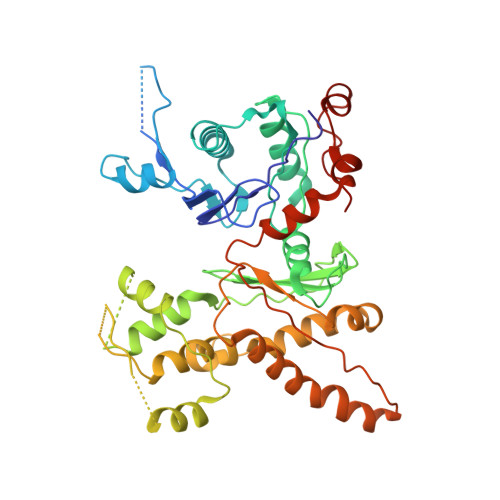
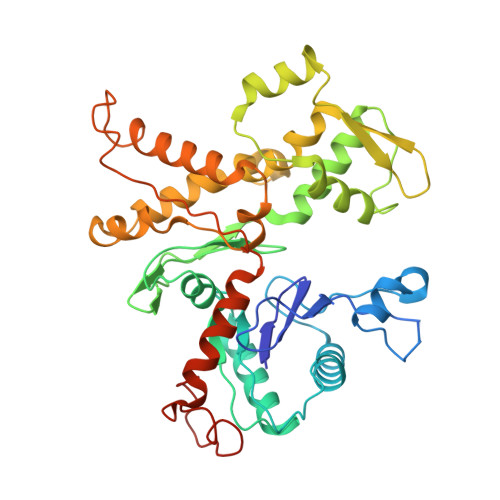
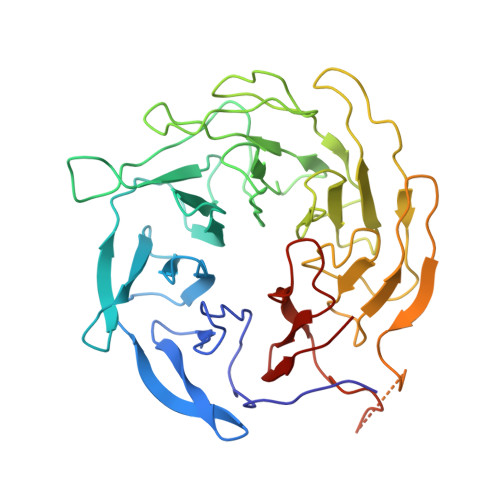
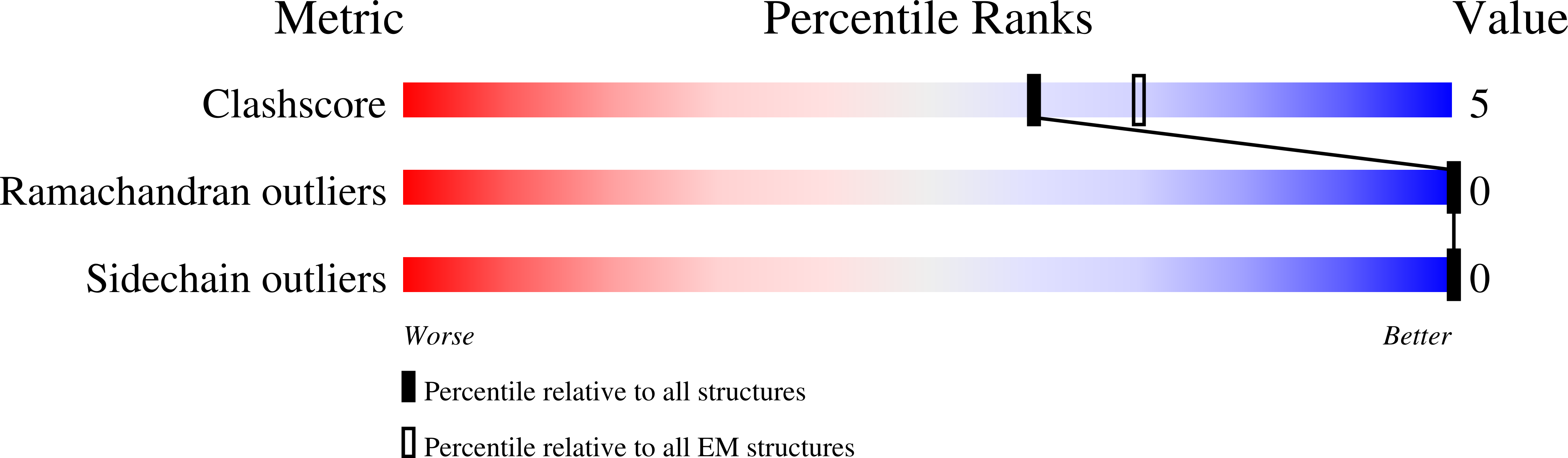Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Shaaban, M., Chowdhury, S., Nolen, B.J.(2020) Nat Struct Mol Biol 27: 1009-1016
- PubMed: 32839613
- DOI: https://doi.org/10.1038/s41594-020-0481-x
- Primary Citation of Related Structures:
6W17, 6W18 - PubMed Abstract:
Arp2/3 complex, a crucial actin filament nucleator, undergoes structural rearrangements during activation by nucleation-promoting factors (NPFs). However, the conformational pathway leading to the nucleation-competent state is unclear due to lack of high-resolution structures of the activated state. Here we report a ~3.9 Å resolution cryo-EM structure of activated Schizosaccharomyces pombe Arp2/3 complex bound to the S. pombe NPF Dip1 and attached to the end of the nucleated actin filament. The structure reveals global and local conformational changes that allow the two actin-related proteins in Arp2/3 complex to mimic a filamentous actin dimer and template nucleation. Activation occurs through a clamp-twisting mechanism, in which Dip1 forces two core subunits in Arp2/3 complex to pivot around one another, shifting half of the complex into a new activated position. By showing how Dip1 stimulates activation, the structure reveals how NPFs can activate Arp2/3 complex in diverse cellular processes.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY, USA.