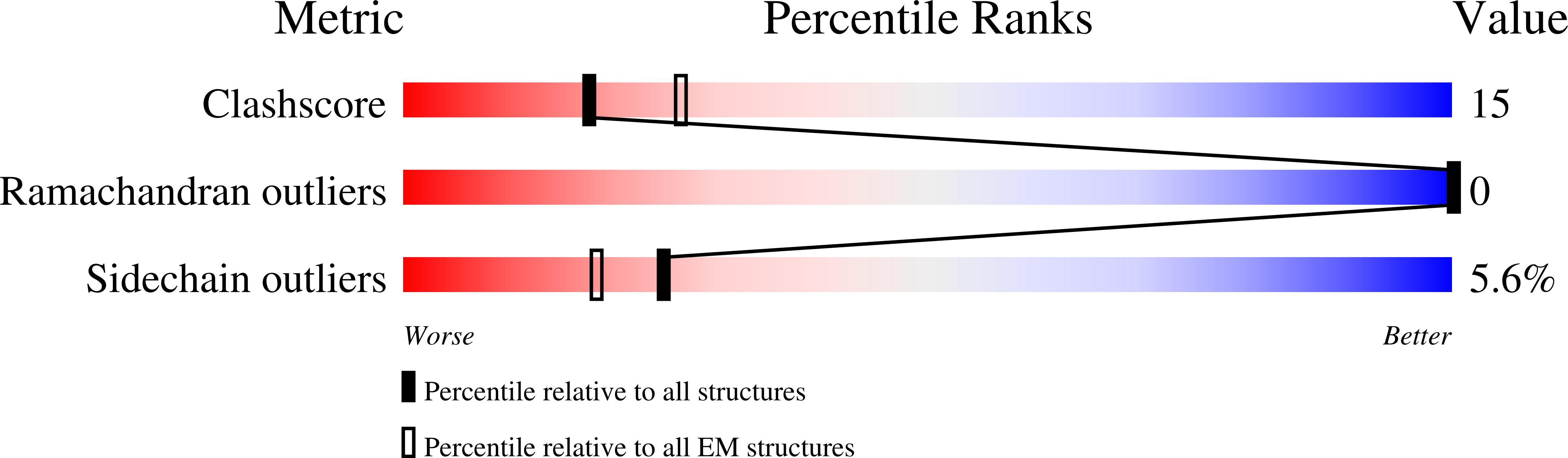Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Kuo, Y.C., Chen, H., Shang, G., Uchikawa, E., Tian, H., Bai, X.C., Zhang, X.(2020) Nat Commun 11: 1953-1953
- PubMed: 32327662
- DOI: https://doi.org/10.1038/s41467-020-15862-0
- Primary Citation of Related Structures:
6VXK - PubMed Abstract:
Plexins are receptors for semaphorins that transduce signals for regulating neuronal development and other processes. Plexins are single-pass transmembrane proteins with multiple domains in both the extracellular and intracellular regions. Semaphorin activates plexin by binding to its extracellular N-terminal Sema domain, inducing the active dimer of the plexin intracellular region. The mechanism underlying this activation process of plexin is incompletely understood. We present cryo-electron microscopic structure of full-length human PlexinC1 in complex with the viral semaphorin mimic A39R. The structure shows that A39R induces a specific dimer of PlexinC1 where the membrane-proximal domains from the two PlexinC1 protomers are placed close to each other, poised to promote the active dimer of the intracellular region. This configuration is imposed by a distinct conformation of the PlexinC1 extracellular region, stabilized by inter-domain interactions among the Sema and membrane-proximal domains. Our mutational analyses support the critical role of this conformation in PlexinC1 activation.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX, USA.