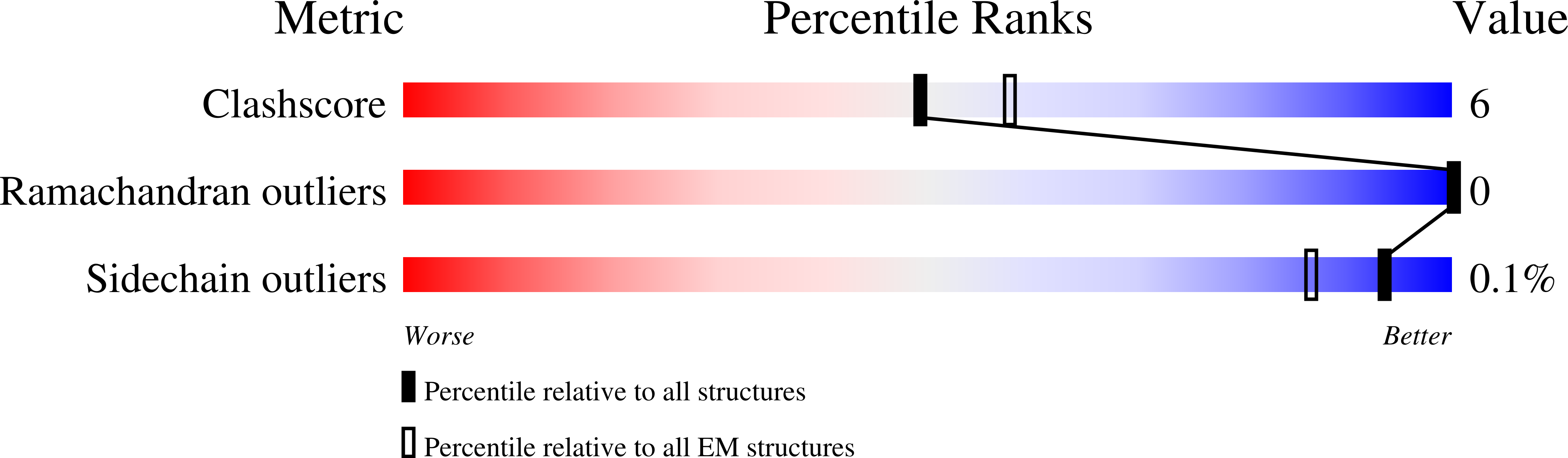Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition.
Gorman, J., Chuang, G.Y., Lai, Y.T., Shen, C.H., Boyington, J.C., Druz, A., Geng, H., Louder, M.K., McKee, K., Rawi, R., Verardi, R., Yang, Y., Zhang, B., Doria-Rose, N.A., Lin, B., Moore, P.L., Morris, L., Shapiro, L., Mascola, J.R., Kwong, P.D.(2020) Cell Rep 31: 107488-107488
- PubMed: 32268107
- DOI: https://doi.org/10.1016/j.celrep.2020.03.052
- Primary Citation of Related Structures:
6VRW, 6VTT - PubMed Abstract:
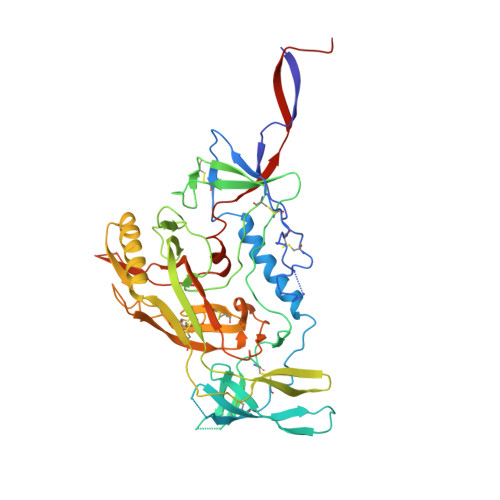
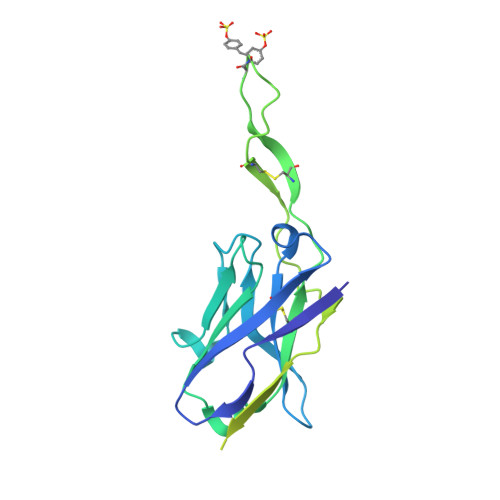
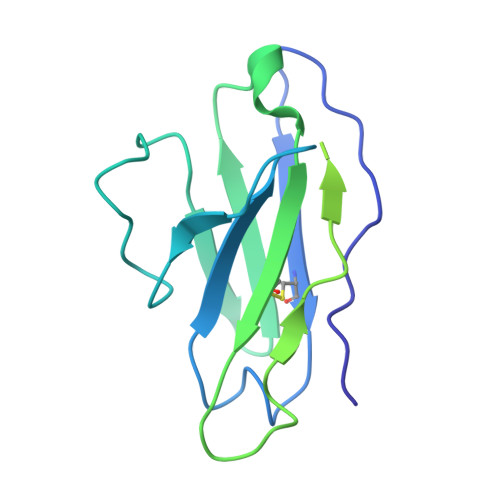
Antibodies targeting the V1V2 apex of the HIV-1 envelope (Env) trimer comprise one of the most commonly elicited categories of broadly neutralizing antibodies. Structures of these antibodies indicate diverse modes of Env recognition typified by antibodies of the PG9 class and the PGT145 class. The mode of recognition, however, has been unclear for the most potent of the V1V2 apex-targeting antibodies, CAP256-VRC26.25 (named for donor-lineage.clone and referred to hereafter as VRC26.25). Here, we determine the cryoelectron microscopy structure at 3.7 Å resolution of the antigen-binding fragment of VRC26.25 in complex with the Env trimer thought to have initiated the lineage. The 36-residue protruding loop of VRC26.25 displays recognition incorporating both strand-C interactions similar to the PG9 class and V1V2 apex insertion similar to the PGT145 class. Structural elements of separate antibody classes can thus intermingle to form a "combined" class, which in this case yields an antibody of extraordinary potency.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.