Engineering Protease-Resistant Peptides to Inhibit Human Parainfluenza Viral Respiratory Infection.
Outlaw, V.K., Cheloha, R.W., Jurgens, E.M., Bovier, F.T., Zhu, Y., Kreitler, D.F., Harder, O., Niewiesk, S., Porotto, M., Gellman, S.H., Moscona, A.(2021) J Am Chem Soc 143: 5958-5966
- PubMed: 33825470
- DOI: https://doi.org/10.1021/jacs.1c01565
- Primary Citation of Related Structures:
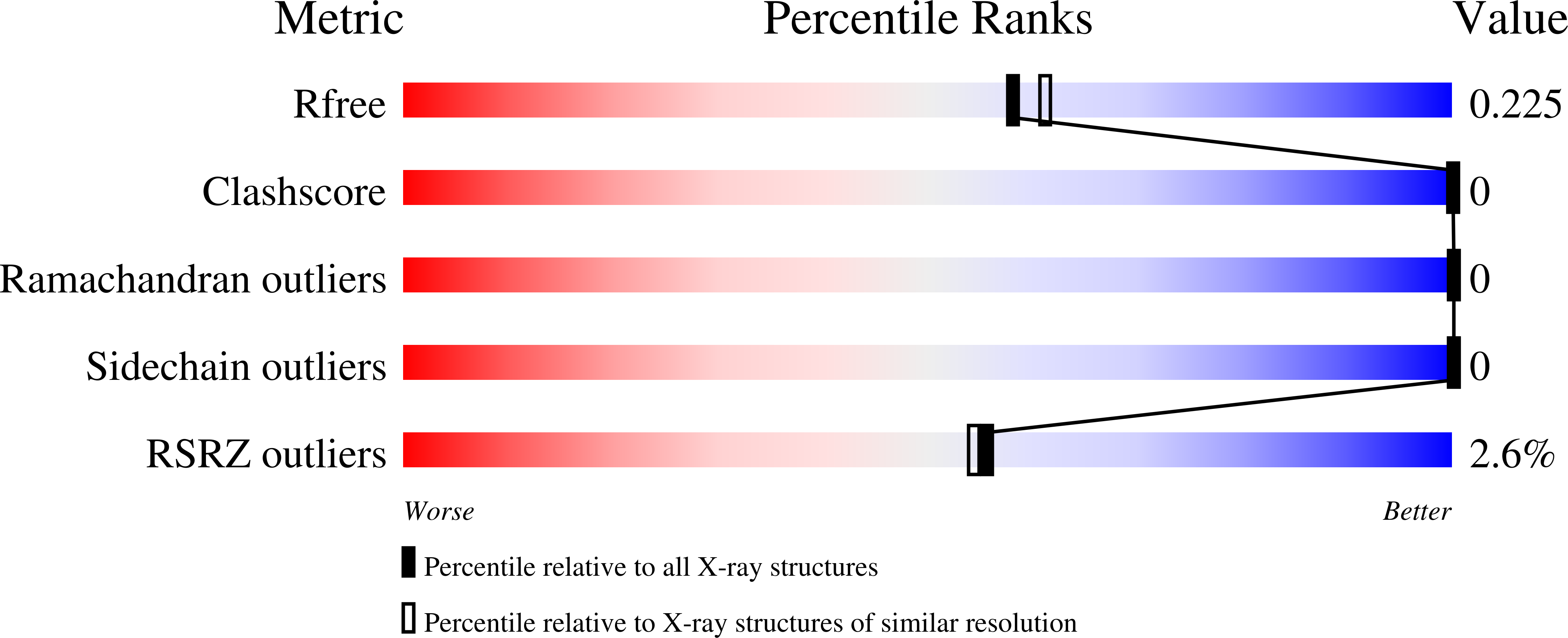
6VJO - PubMed Abstract:
The lower respiratory tract infections affecting children worldwide are in large part caused by the parainfluenza viruses (HPIVs), particularly HPIV3, along with human metapneumovirus and respiratory syncytial virus, enveloped negative-strand RNA viruses. There are no vaccines for these important human pathogens, and existing treatments have limited or no efficacy. Infection by HPIV is initiated by viral glycoprotein-mediated fusion between viral and host cell membranes. A viral fusion protein (F), once activated in proximity to a target cell, undergoes a series of conformational changes that first extend the trimer subunits to allow insertion of the hydrophobic domains into the target cell membrane and then refold the trimer into a stable postfusion state, driving the merger of the viral and host cell membranes. Lipopeptides derived from the C-terminal heptad repeat (HRC) domain of HPIV3 F inhibit infection by interfering with the structural transitions of the trimeric F assembly. Clinical application of this strategy, however, requires improving the in vivo stability of antiviral peptides. We show that the HRC peptide backbone can be modified via partial replacement of α-amino acid residues with β-amino acid residues to generate α/β-peptides that retain antiviral activity but are poor protease substrates. Relative to a conventional α-lipopeptide, our best α/β-lipopeptide exhibits improved persistence in vivo and improved anti-HPIV3 antiviral activity in animals.
Organizational Affiliation:
Department of Chemistry, University of Wisconsin, Madison, Wisconsin 53706, United States.