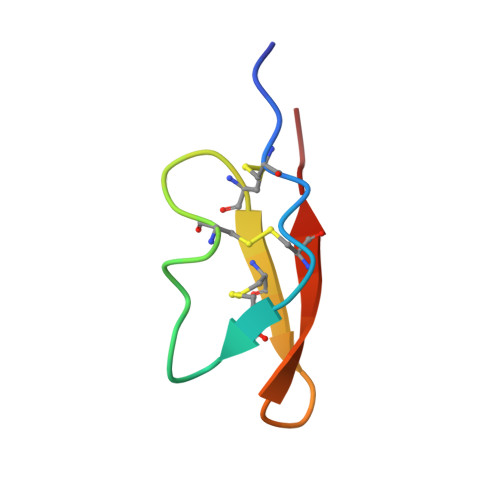
Neurotoxic peptides from the venom of the giant Australian stinging tree.
Gilding, E.K., Jami, S., Deuis, J.R., Israel, M.R., Harvey, P.J., Poth, A.G., Rehm, F.B.H., Stow, J.L., Robinson, S.D., Yap, K., Brown, D.L., Hamilton, B.R., Andersson, D., Craik, D.J., Vetter, I., Durek, T.(2020) Sci Adv 6
- PubMed: 32938666
- DOI: https://doi.org/10.1126/sciadv.abb8828
- Primary Citation of Related Structures:
6VH8 - PubMed Abstract:
Stinging trees from Australasia produce remarkably persistent and painful stings upon contact of their stiff epidermal hairs, called trichomes, with mammalian skin. Dendrocnide -induced acute pain typically lasts for several hours, and intermittent painful flares can persist for days and weeks. Pharmacological activity has been attributed to small-molecule neurotransmitters and inflammatory mediators, but these compounds alone cannot explain the observed sensory effects. We show here that the venoms of Australian Dendrocnide species contain heretofore unknown pain-inducing peptides that potently activate mouse sensory neurons and delay inactivation of voltage-gated sodium channels. These neurotoxins localize specifically to the stinging hairs and are miniproteins of 4 kDa, whose 3D structure is stabilized in an inhibitory cystine knot motif, a characteristic shared with neurotoxins found in spider and cone snail venoms. Our results provide an intriguing example of inter-kingdom convergent evolution of animal and plant venoms with shared modes of delivery, molecular structure, and pharmacology.
Organizational Affiliation:
Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD 4072, Australia.