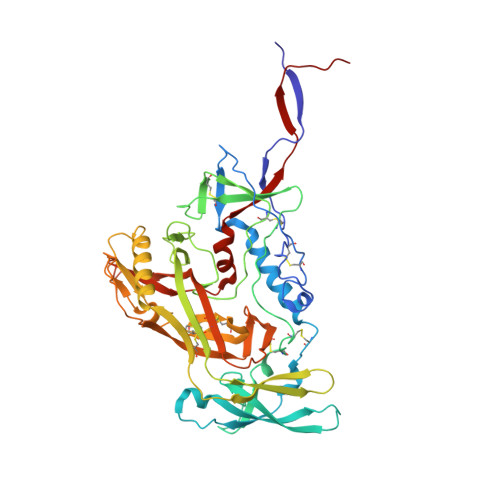
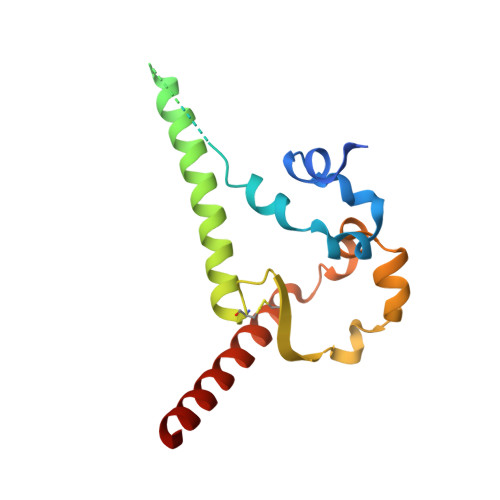
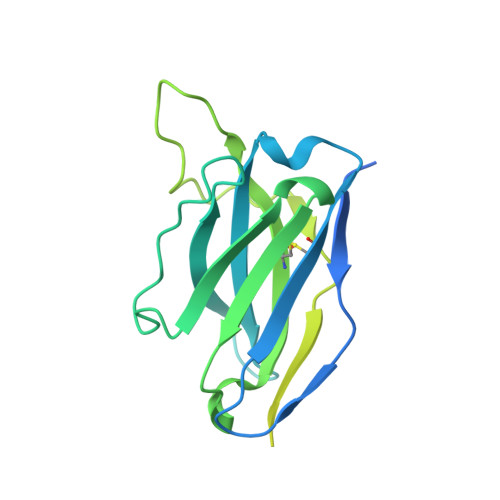
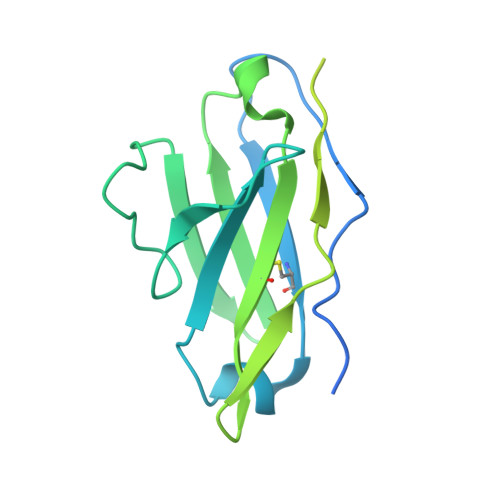
Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Saunders, K.O., Wiehe, K., Tian, M., Acharya, P., Bradley, T., Alam, S.M., Go, E.P., Scearce, R., Sutherland, L., Henderson, R., Hsu, A.L., Borgnia, M.J., Chen, H., Lu, X., Wu, N.R., Watts, B., Jiang, C., Easterhoff, D., Cheng, H.L., McGovern, K., Waddicor, P., Chapdelaine-Williams, A., Eaton, A., Zhang, J., Rountree, W., Verkoczy, L., Tomai, M., Lewis, M.G., Desaire, H.R., Edwards, R.J., Cain, D.W., Bonsignori, M., Montefiori, D., Alt, F.W., Haynes, B.F.(2019) Science 366
- PubMed: 31806786
- DOI: https://doi.org/10.1126/science.aay7199
- Primary Citation of Related Structures:
6UM5, 6UM6, 6UM7 - PubMed Abstract:
A major goal of HIV-1 vaccine development is the design of immunogens that induce broadly neutralizing antibodies (bnAbs). However, vaccination of humans has not resulted in the induction of affinity-matured and potent HIV-1 bnAbs. To devise effective vaccine strategies, we previously determined the maturation pathway of select HIV-1 bnAbs from acute infection through neutralizing antibody development. During their evolution, bnAbs acquire an abundance of improbable amino acid substitutions as a result of nucleotide mutations at variable region sequences rarely targeted by activation-induced cytidine deaminase, the enzyme responsible for antibody mutation. A subset of improbable mutations is essential for broad neutralization activity, and their acquisition represents a key roadblock to bnAb development.
Organizational Affiliation:
Human Vaccine Institute and Department of Surgery, Duke University School of Medicine, Durham, NC 27710, USA. kevin.saunders@duke.edu barton.haynes@duke.edu alt@enders.tch.harvard.edu.