Discovery of Small-Molecule Antagonists of the PWWP Domain of NSD2.
Ferreira de Freitas, R., Liu, Y., Szewczyk, M.M., Mehta, N., Li, F., McLeod, D., Zepeda-Velazquez, C., Dilworth, D., Hanley, R.P., Gibson, E., Brown, P.J., Al-Awar, R., James, L.I., Arrowsmith, C.H., Barsyte-Lovejoy, D., Min, J., Vedadi, M., Schapira, M., Allali-Hassani, A.(2021) J Med Chem 64: 1584-1592
- PubMed: 33522809
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01768
- Primary Citation of Related Structures:
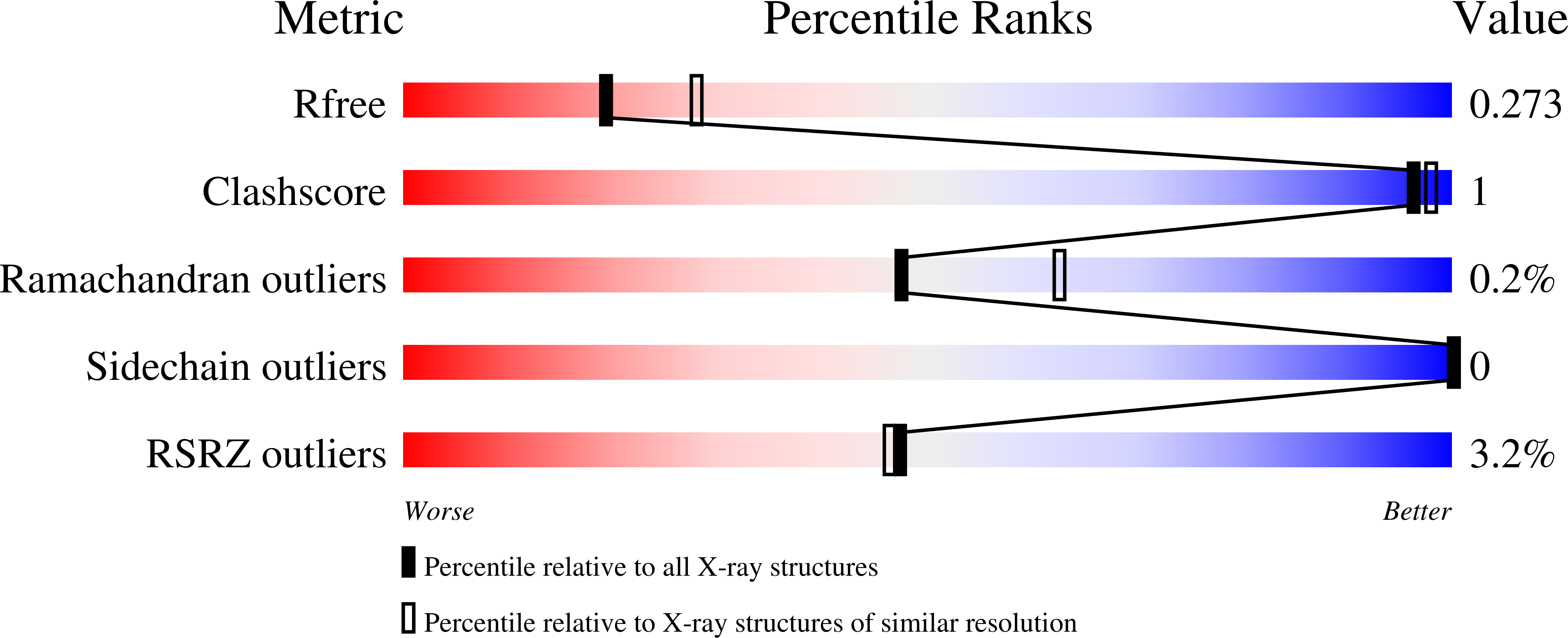
6UE6 - PubMed Abstract:
Increased activity of the lysine methyltransferase NSD2 driven by translocation and activating mutations is associated with multiple myeloma and acute lymphoblastic leukemia, but no NSD2-targeting chemical probe has been reported to date. Here, we present the first antagonists that block the protein-protein interaction between the N-terminal PWWP domain of NSD2 and H3K36me2. Using virtual screening and experimental validation, we identified the small-molecule antagonist 3f , which binds to the NSD2-PWWP1 domain with a K d of 3.4 μM and abrogates histone H3K36me2 binding to the PWWP1 domain in cells. This study establishes an alternative approach to targeting NSD2 and provides a small-molecule antagonist that can be further optimized into a chemical probe to better understand the cellular function of this protein.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7, Canada.