Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Cho, J.H., Zhao, B., Shi, J., Savage, N., Shen, Q., Byrnes, J., Yang, L., Hwang, W., Li, P.(2020) Proc Natl Acad Sci U S A 117: 6550-6558
- PubMed: 32152123
- DOI: https://doi.org/10.1073/pnas.1920582117
- Primary Citation of Related Structures:
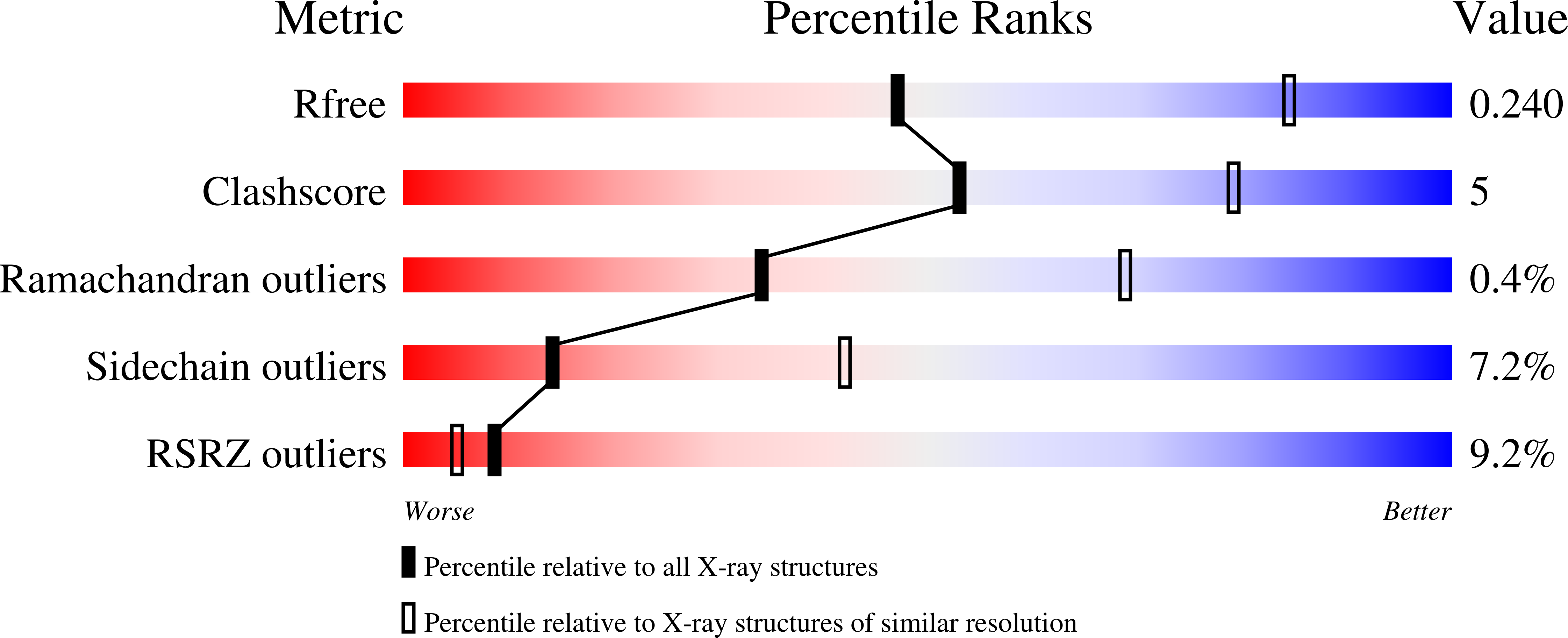
6OX7, 6U28 - PubMed Abstract:
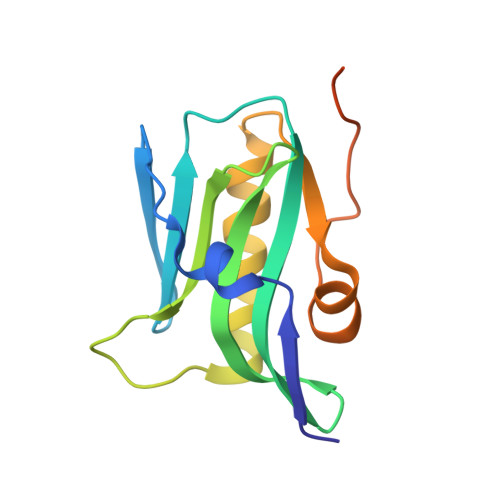
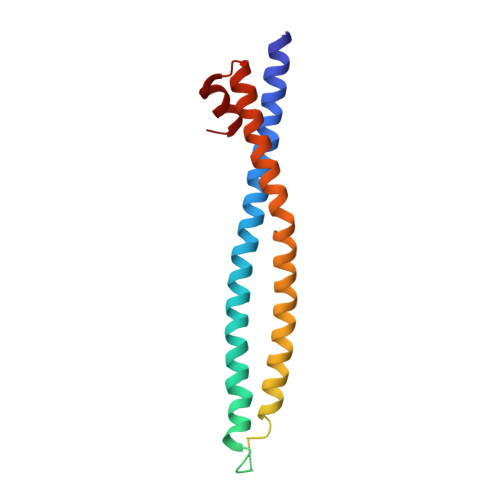
The 1918 influenza A virus (IAV) caused the most severe flu pandemic in recorded human history. Nonstructural protein 1 (NS1) is an important virulence factor of the 1918 IAV. NS1 antagonizes host defense mechanisms through interactions with multiple host factors. One pathway by which NS1 increases virulence is through the activation of phosphoinositide 3-kinase (PI3K) by binding to its p85β subunit. Here we present the mechanism underlying the molecular recognition of the p85β subunit by 1918 NS1. Using X-ray crystallography, we determine the structure of 1918 NS1 complexed with p85β of human PI3K. We find that the 1918 NS1 effector domain (1918 NS1 ED ) undergoes a conformational change to bind p85β. Using NMR relaxation dispersion and molecular dynamics simulation, we identify that free 1918 NS1 ED exists in a dynamic equilibrium between p85β-binding-competent and -incompetent conformations in the submillisecond timescale. Moreover, we discover that NS1 ED proteins of 1918 (H1N1) and Udorn (H3N2) strains exhibit drastically different conformational dynamics and binding kinetics to p85β. These results provide evidence of strain-dependent conformational dynamics of NS1. Using kinetic modeling based on the experimental data, we demonstrate that 1918 NS1 ED can result in the faster hijacking of p85β compared to Ud NS1 ED , although the former has a lower affinity to p85β than the latter. Our results suggest that the difference in binding kinetics may impact the competition with cellular antiviral responses for the activation of PI3K. We anticipate that our findings will increase the understanding of the strain-dependent behaviors of influenza NS1 proteins.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843; jaehyuncho@tamu.edu.