Two independent routes of post-translational chemistry in fluorescent protein FusionRed.
Muslinkina, L., Pletnev, V.Z., Pletneva, N.V., Ruchkin, D.A., Kolesov, D.V., Bogdanov, A.M., Kost, L.A., Rakitina, T.V., Agapova, Y.K., Shemyakina, I.I., Chudakov, D.M., Pletnev, S.(2020) Int J Biol Macromol 155: 551-559
- PubMed: 32243936
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.03.244
- Primary Citation of Related Structures:
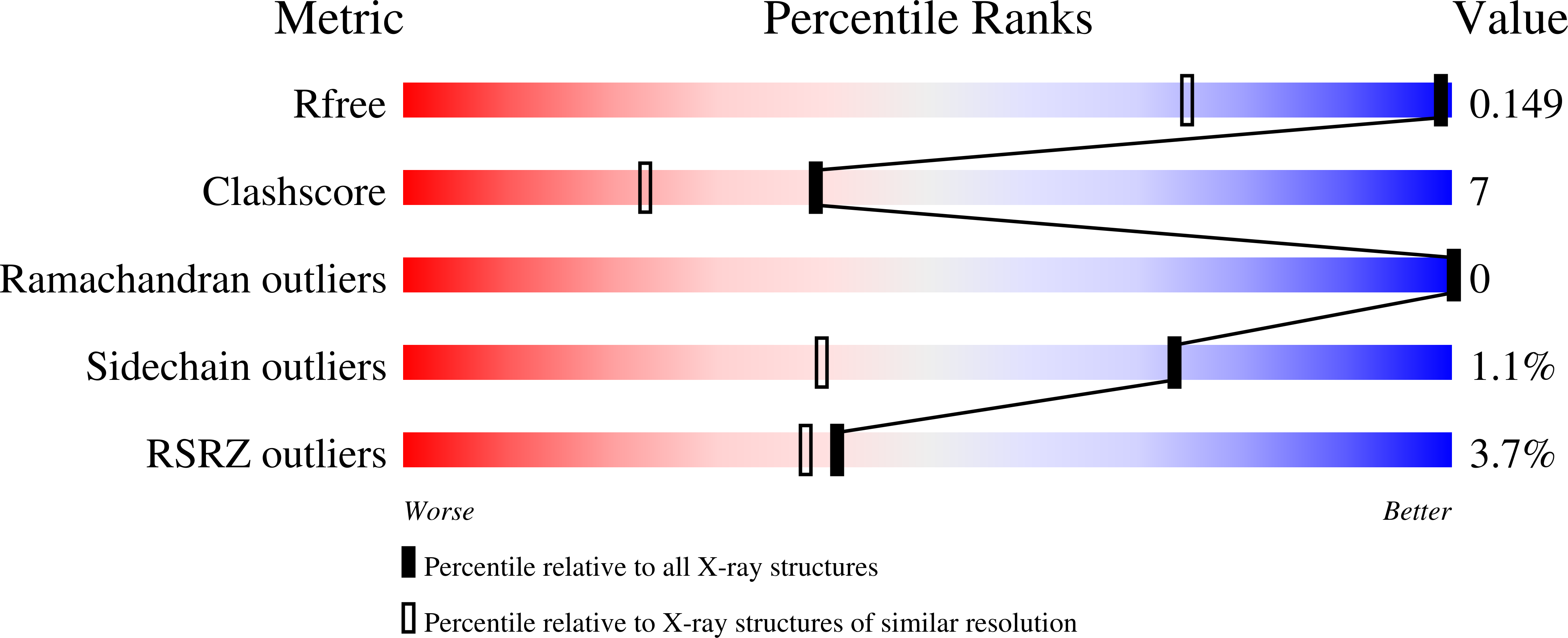
6U1A - PubMed Abstract:
The crystal structure of monomeric red fluorescent protein FusionRed (λ ex /λ em 580/608 mn) has been determined at 1.09 Å resolution and revealed two alternative routes of post-translational chemistry, resulting in distinctly different products. The refinement occupancies suggest the 60:40 ratio of the mature Met63-Tyr64-Gly65 chromophore and uncyclized chromophore-forming tripeptide with the protein backbone cleaved between Met63 and the preceding Phe62 and oxidized Cα-Cβ bond of Tyr64. We analyzed the structures of FusionRed and several related red fluorescent proteins, identified structural elements causing hydrolysis of the peptide bond, and verified their impact by single point mutagenesis. These findings advance the understanding of the post-translational chemistry of GFP-like fluorescent proteins beyond the canonical cyclization-dehydration-oxidation mechanism. They also show that impaired cyclization does not prevent chromophore-forming tripeptide from further transformations enabled by the same set of catalytic residues. Our mutagenesis efforts resulted in inhibition of the peptide backbone cleavage, and a FusionRed variant with ~30% improved effective brightness.
Organizational Affiliation:
Basic Science Program, Frederick National Laboratory for Cancer Research, Argonne, IL 60439, USA.