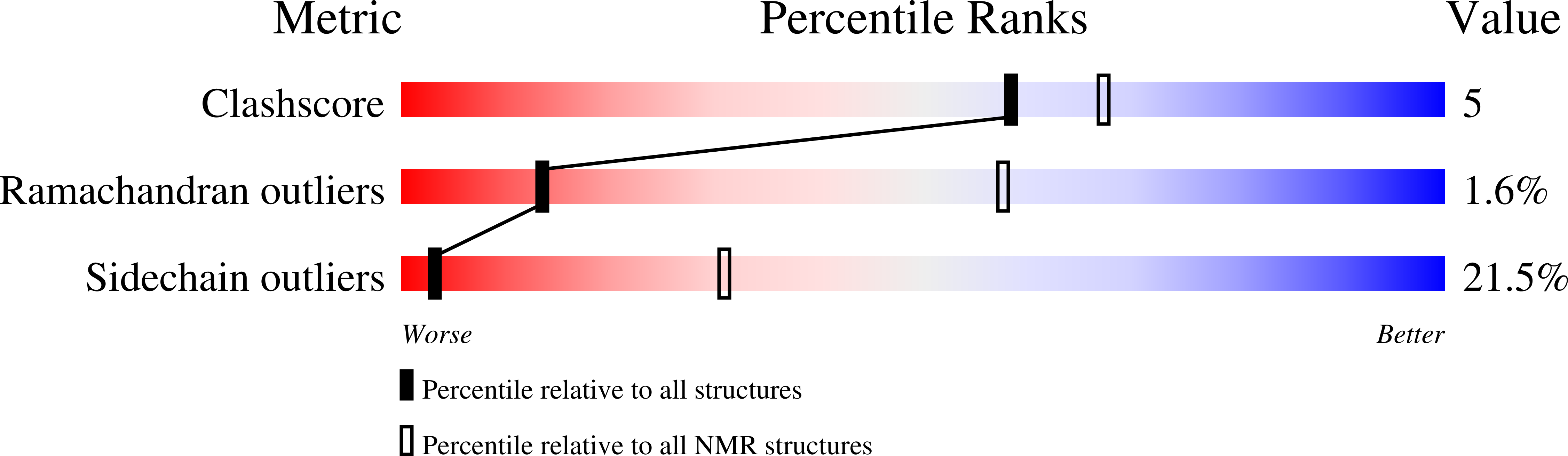Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Czajlik, A., Holzknecht, J., Galgoczy, L., Toth, L., Poor, P., Ordog, A., Varadi, G., Kuhbacher, A., Borics, A., Toth, G.K., Marx, F., Batta, G.(2021) Int J Mol Sci 22
- PubMed: 33504082
- DOI: https://doi.org/10.3390/ijms22031183
- Primary Citation of Related Structures:
6TRM - PubMed Abstract:
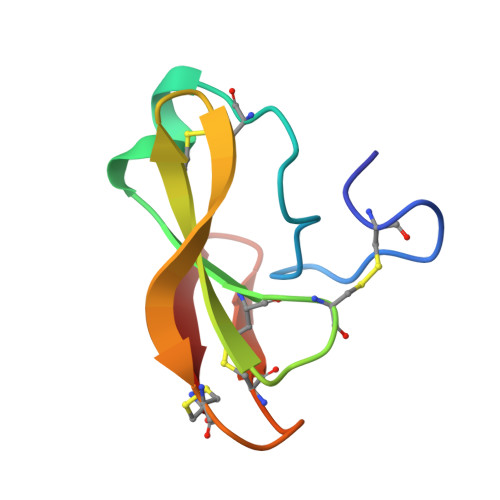
The genome of Penicillium chrysogenum Q176 contains a gene coding for the 88-amino-acid (aa)-long glycine- and cysteine-rich P. chrysogenum antifungal protein C (PAFC). After maturation, the secreted antifungal miniprotein (MP) comprises 64 aa and shares 80% aa identity with the bubble protein (BP) from Penicillium brevicompactum , which has a published X-ray structure. Our team expressed isotope ( 15 N, 13 C)-labeled, recombinant PAFC in high yields, which allowed us to determine the solution structure and molecular dynamics by nuclear magnetic resonance (NMR) experiments. The primary structure of PAFC is dominated by 14 glycines, and therefore, whether the four disulfide bonds can stabilize the fold is challenging. Indeed, unlike the few published solution structures of other antifungal MPs from filamentous ascomycetes, the NMR data indicate that PAFC has shorter secondary structure elements and lacks the typical β-barrel structure, though it has a positively charged cavity and a hydrophobic core around the disulfide bonds. Some parts within the two putative γ-core motifs exhibited enhanced dynamics according to a new disorder index presentation of 15 N-NMR relaxation data. Furthermore, we also provided a more detailed insight into the antifungal spectrum of PAFC, with specific emphasis on fungal plant pathogens. Our results suggest that PAFC could be an effective candidate for the development of new antifungal strategies in agriculture.
Organizational Affiliation:
Department of Organic Chemistry, Faculty of Science and Technology, University of Debrecen, H-4032 Debrecen, Hungary.