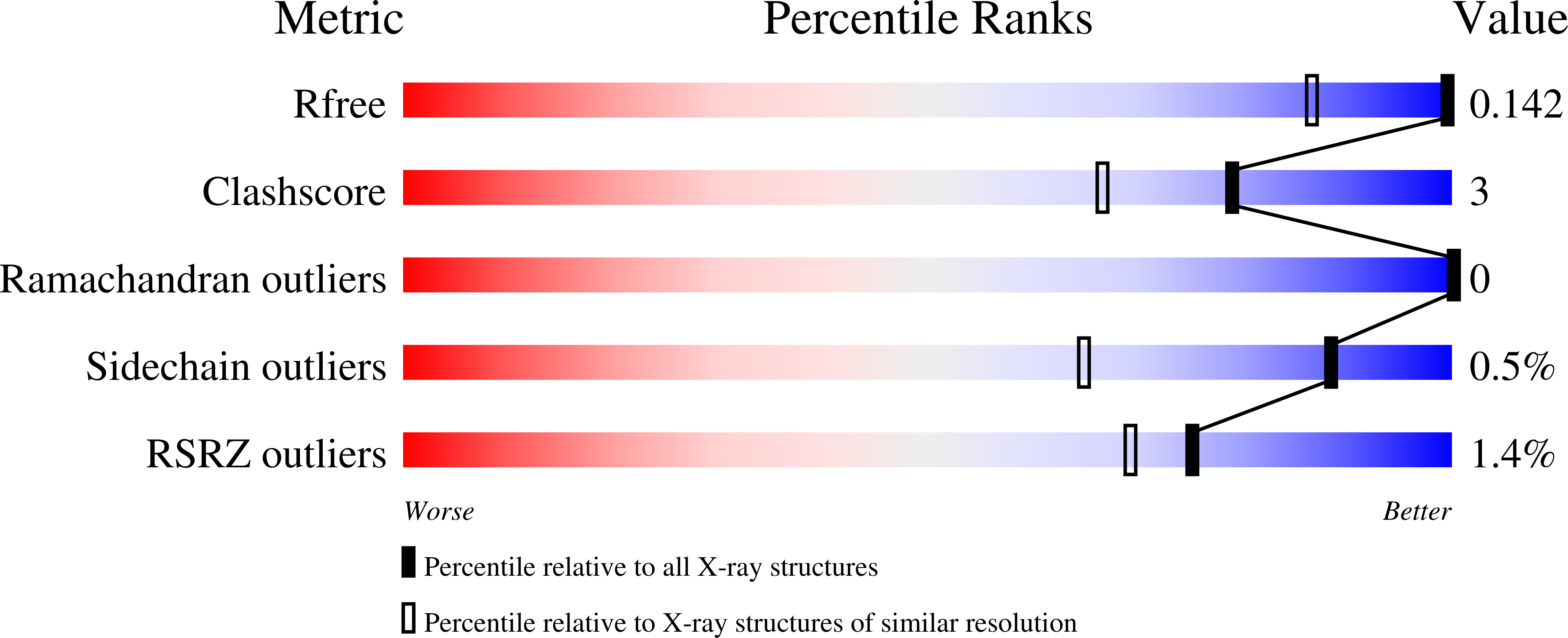
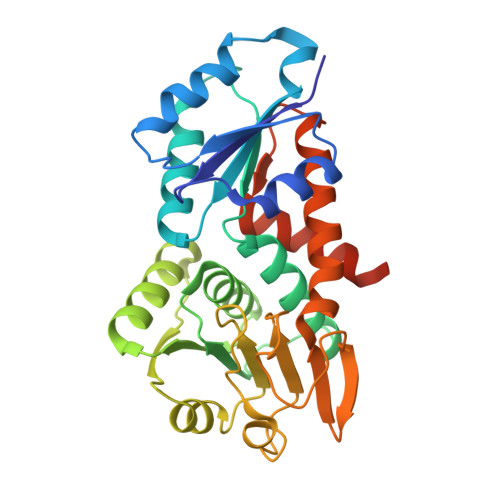
The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
Huang, G., Wagner, T., Demmer, U., Warkentin, E., Ermler, U., Shima, S.(2020) J Mol Biol 432: 2042-2054
- PubMed: 32061937
- DOI: https://doi.org/10.1016/j.jmb.2020.01.042
- Primary Citation of Related Structures:
6TGE, 6TLK, 6TM3, 6YK9, 6YKA, 6YKB - PubMed Abstract:
NADP-dependent methylene-tetrahydromethanopterin (methylene-H 4 MPT) dehydrogenase (MtdA) catalyzes the reversible dehydrogenation of methylene-H 4 MPT to form methenyl-H 4 MPT + by using NADP + as a hydride acceptor. This hydride transfer reaction is involved in the oxidative metabolism from formaldehyde to CO 2 in methylotrophic and methanotrophic bacteria. Here, we report on the crystal structures of the ternary MtdA-substrate complexes from Methylorubrum extorquens AM1 obtained in open and closed forms. Their conversion is accomplished by opening/closing the active site cleft via a 15° rotation of the NADP, relative to the pterin domain. The 1.08 Å structure of the closed and active enzyme-NADP-methylene-H 4 MPT complex allows a detailed geometric analysis of the bulky substrates and a precise prediction of the hydride trajectory. Upon domain closure, the bulky substrate rings become compressed resulting in a tilt of the imidazolidine group of methylene-H 4 MPT that optimizes the geometry for hydride transfer. An additional 1.5 Å structure of MtdA in complex with the nonreactive NADP + and methenyl-H 4 MPT + revealed an extremely short distance between nicotinamide-C4 and imidazoline-C14a of 2.5 Å, which demonstrates the strong pressure imposed. The pterin-imidazolidine-phenyl butterfly angle of methylene-H 4 MPT bound to MtdA is smaller than that in the enzyme-free state but is similar to that in H 2 - and F 420 -dependent methylene-H 4 MPT dehydrogenases. The concept of compression-driven hydride transfer including quantum mechanical hydrogen tunneling effects, which are established for flavin- and NADP-dependent enzymes, can be expanded to hydride-transferring H 4 MPT-dependent enzymes.
Organizational Affiliation:
Max-Planck-Institut für terrestrische Mikrobiologie, Karl-von-Frisch Straße 10, 35043, Marburg, Germany.