Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Gregory, K.S., Liu, S.M., Acharya, K.R.(2020) FEBS Open Bio 10: 298-305
- PubMed: 31945264
- DOI: https://doi.org/10.1002/2211-5463.12790
- Primary Citation of Related Structures:
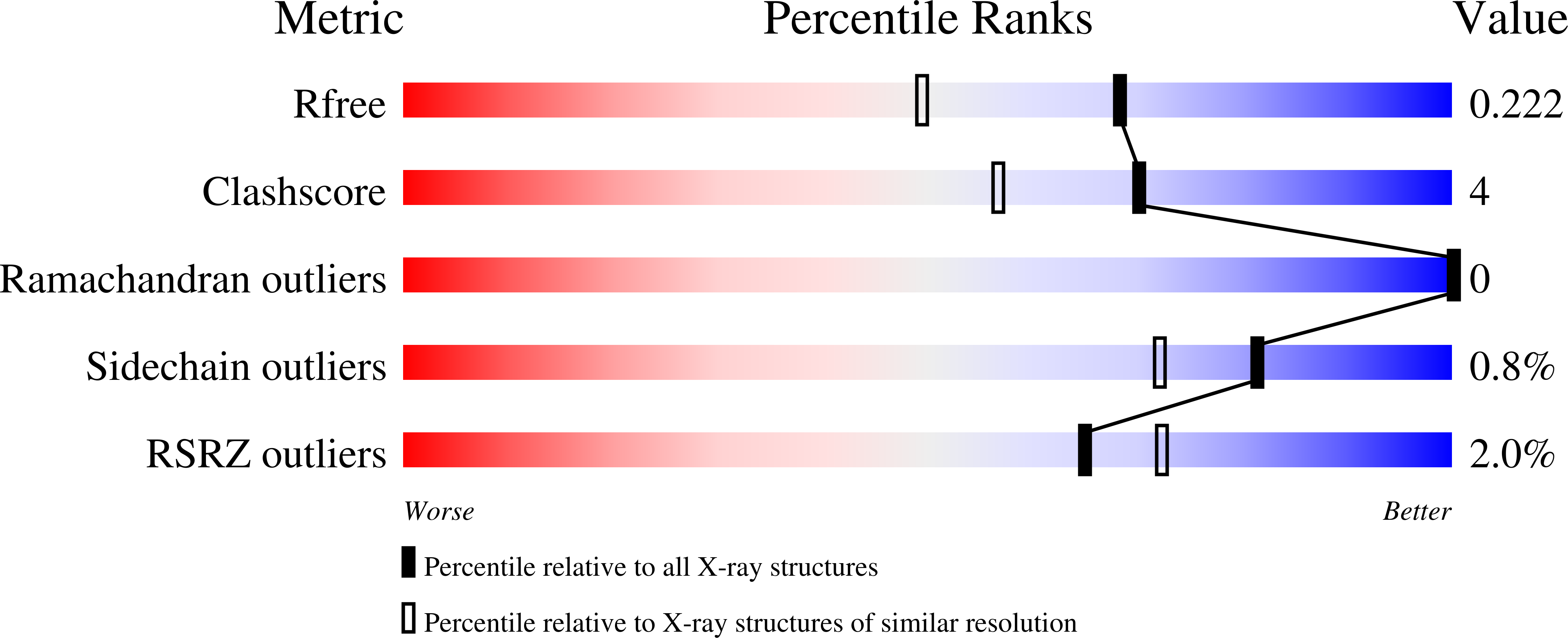
6THY - PubMed Abstract:
Botulinum neurotoxins (BoNTs) are one of the most toxic proteins known to humans. Their molecular structure is comprised of three essential domains-a cell binding domain (H C ), translocation domain and catalytic domain (light chain) . The H C domain facilitates the highly specific binding of BoNTs to the neuronal membrane via a dual-receptor complex involving a protein receptor and a ganglioside. Variation in activity/toxicity across subtypes of serotype A has been attributed to changes in protein and ganglioside interactions, and their implications are important in the design of novel BoNT-based therapeutics. Here, we present the structure of BoNT/A3 cell binding domain (H C /A3) in complex with the ganglioside GD1a at 1.75 Å resolution. The structure revealed that six residues interact with the three outermost monosaccharides of GD1a through several key hydrogen bonding interactions. A detailed comparison of structures of H C /A3 with H C /A1 revealed subtle conformational differences at the ganglioside binding site upon carbohydrate binding.
Organizational Affiliation:
Department of Biology and Biochemistry, Claverton Down, University of Bath, UK.