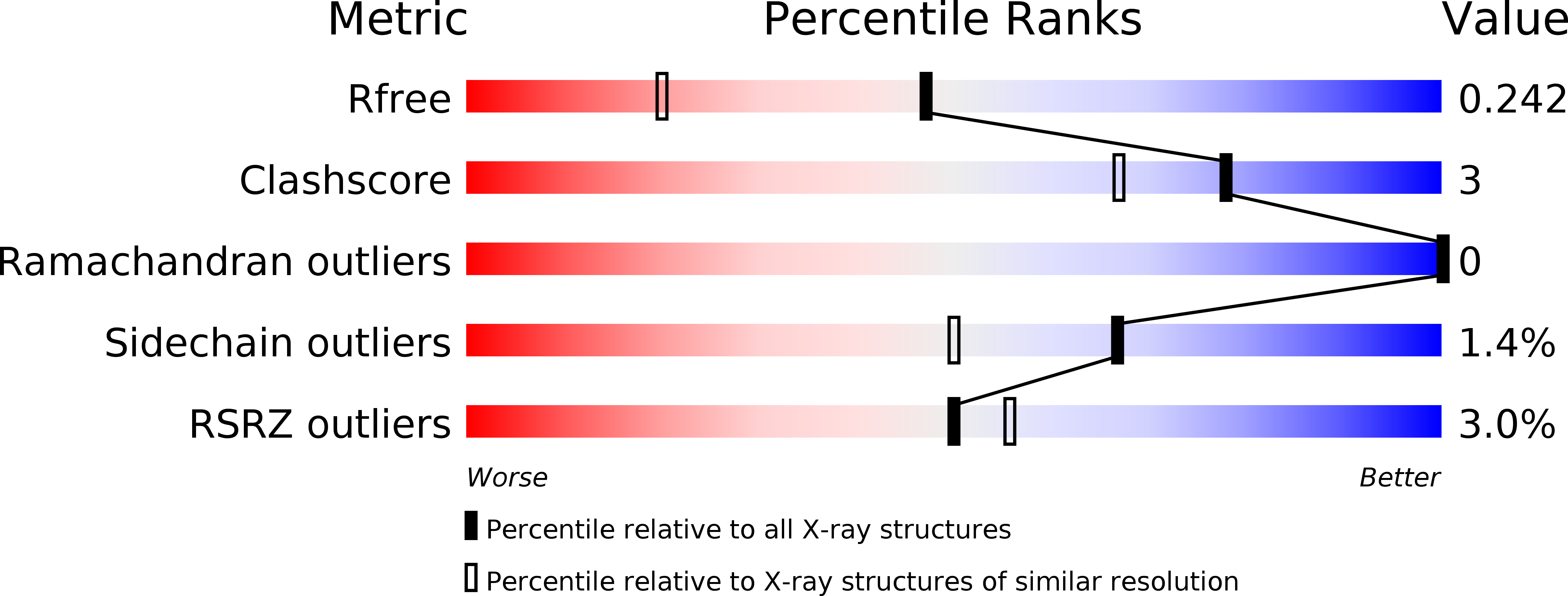
Targeting a critical step in fungal hexosamine biosynthesis.
Lockhart, D.E.A., Stanley, M., Raimi, O.G., Robinson, D.A., Boldovjakova, D., Squair, D.R., Ferenbach, A.T., Fang, W., van Aalten, D.M.F.(2020) J Biol Chem 295: 8678-8691
- PubMed: 32341126
- DOI: https://doi.org/10.1074/jbc.RA120.012985
- Primary Citation of Related Structures:
6TDF, 6TDG, 6TDH - PubMed Abstract:
Aspergillus fumigatus is a human opportunistic fungal pathogen whose cell wall protects it from the extracellular environment including host defenses. Chitin, an essential component of the fungal cell wall, is synthesized from UDP-GlcNAc produced in the hexosamine biosynthetic pathway. As this pathway is critical for fungal cell wall integrity, the hexosamine biosynthesis enzymes represent potential targets of antifungal drugs. Here, we provide genetic and chemical evidence that glucosamine 6-phosphate N -acetyltransferase (Gna1), a key enzyme in this pathway, is an exploitable antifungal drug target. GNA1 deletion resulted in loss of fungal viability and disruption of the cell wall, phenotypes that could be rescued by exogenous GlcNAc, the product of the Gna1 enzyme. In a murine model of aspergillosis, the Δ gna1 mutant strain exhibited attenuated virulence. Using a fragment-based approach, we discovered a small heterocyclic scaffold that binds proximal to the Gna1 active site and can be optimized to a selective submicromolar binder. Taken together, we have provided genetic, structural, and chemical evidence that Gna1 is an antifungal target in A. fumigatus .
Organizational Affiliation:
School of Life Sciences, University of Dundee, Dundee, United Kingdom; Institute of Medical Sciences, Foresterhill, University of Aberdeen, Aberdeen, United Kingdom. Electronic address: deborah.lockhart@abdn.ac.uk.