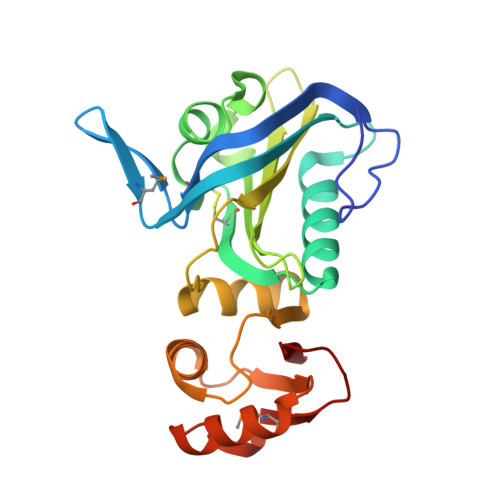
The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Feiler, C.G., Weiss, M.S., Blankenfeldt, W.(2020) Acta Crystallogr F Struct Biol Commun 76: 609-615
- PubMed: 33263573
- DOI: https://doi.org/10.1107/S2053230X20014612
- Primary Citation of Related Structures:
6TD9 - PubMed Abstract:
The crystal structure of the 268-residue periplasmic protein PA1624 from the opportunistic pathogen Pseudomonas aeruginosa PAO1 was determined to high resolution using the Se-SAD method for initial phasing. The protein was found to be monomeric and the structure consists of two domains, domains 1 and 2, comprising residues 24-184 and 185-268, respectively. The fold of these domains could not be predicted even using state-of-the-art prediction methods, and similarity searches revealed only a very distant homology to known structures, namely to Mog1p/PsbP-like and OmpA-like proteins for the N- and C-terminal domains, respectively. Since PA1624 is only present in an important human pathogen, its unique structure and periplasmic location render it a potential drug target. Consequently, the results presented here may open new avenues for the discovery and design of antibacterial drugs.
Organizational Affiliation:
Macromolecular Crystallography (HZB-MX), Helmholtz-Zentrum Berlin, Albert-Einstein-Strasse 15, D-12489 Berlin, Germany.