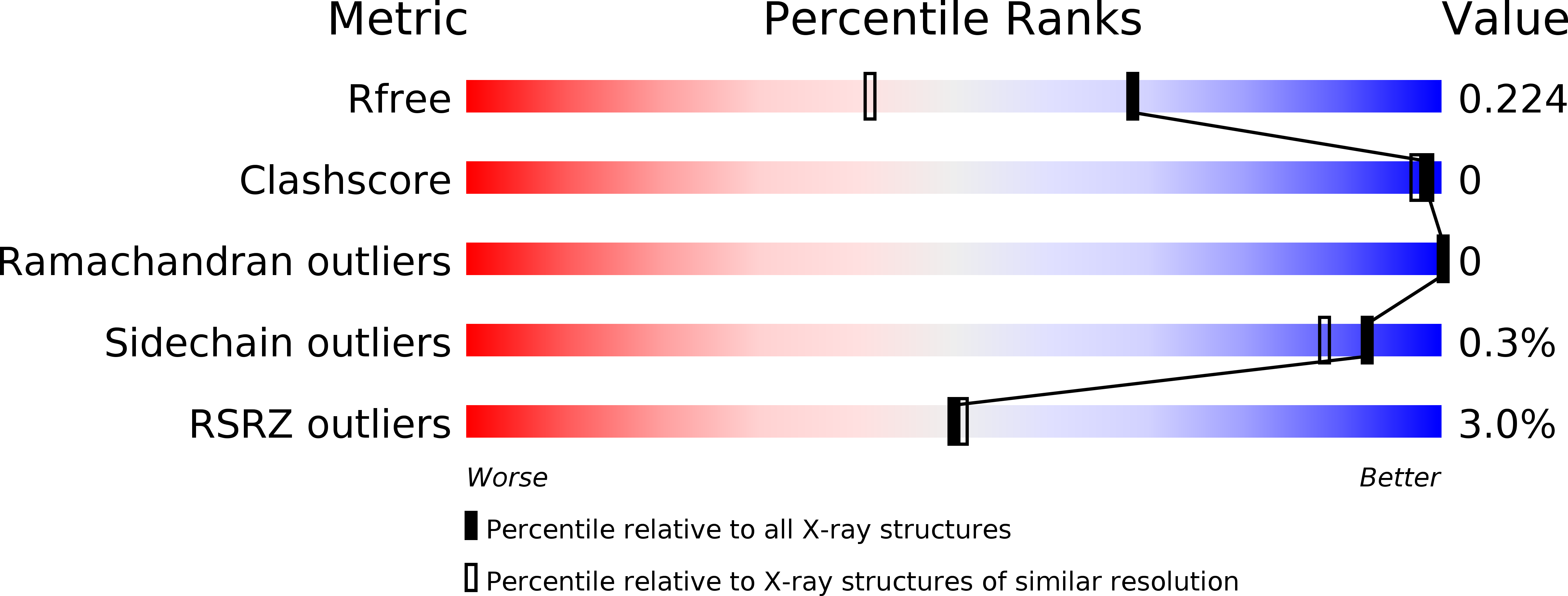
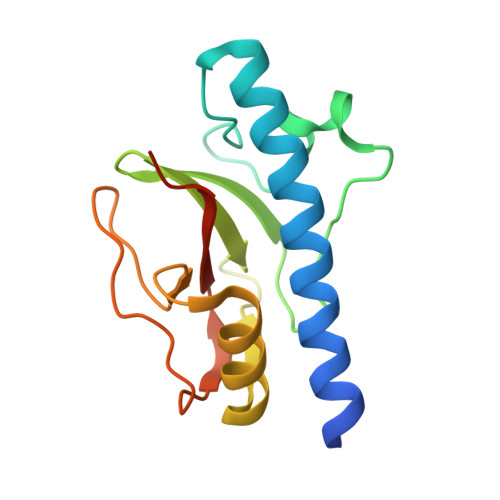
A practical overview of molecular replacement: Clostridioides difficile PilA1, a difficult case study.
Crawshaw, A.D., Basle, A., Salgado, P.S.(2020) Acta Crystallogr D Struct Biol 76: 261-271
- PubMed: 32133990
- DOI: https://doi.org/10.1107/S2059798320000467
- Primary Citation of Related Structures:
6T8S - PubMed Abstract:
Many biologists are now routinely seeking to determine the three-dimensional structures of their proteins of choice, illustrating the importance of this knowledge, but also of the simplification and streamlining of structure-determination processes. Despite the fact that most software packages offer simple pipelines, for the non-expert navigating the outputs and understanding the key aspects can be daunting. Here, the structure determination of the type IV pili (TFP) protein PilA1 from Clostridioides difficile is used to illustrate the different steps involved, the key decision criteria and important considerations when using the most common pipelines and software. Molecular-replacement pipelines within CCP4i2 are presented to illustrate the more commonly used processes. Previous knowledge of the biology and structure of TFP pilins, particularly the presence of a long, N-terminal α-helix required for pilus formation, allowed informed decisions to be made during the structure-determination strategy. The PilA1 structure was finally successfully determined using ARCIMBOLDO and the ab initio MR strategy used is described.
Organizational Affiliation:
Newcastle University Biosciences Institute, Faculty of Medical Sciences, Newcastle University, Newcastle upon Tyne, England.