Crystal structure of the accessory translocation ATPase, SecA2, from Clostridium difficile
Lindic, N., Loboda, J., Usenik, A., Turk, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein translocase subunit SecA 2 | 786 | Clostridioides difficile 630 | Mutation(s): 0 Gene Names: secA2, CD630_27920 | 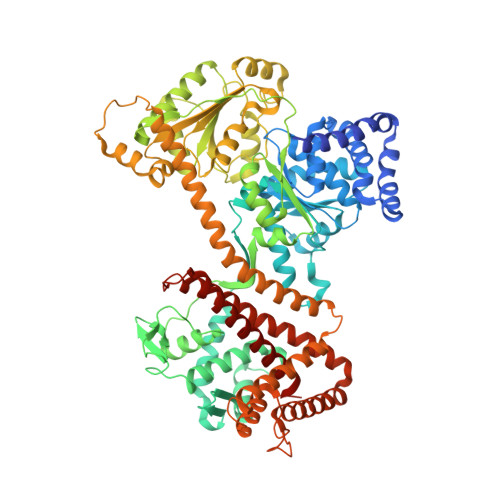 | |
UniProt | |||||
Find proteins for Q183M9 (Clostridioides difficile (strain 630)) Explore Q183M9 Go to UniProtKB: Q183M9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q183M9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 84.99 | α = 90 |
| b = 96.85 | β = 90 |
| c = 114.74 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAIN | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Slovenian Research Agency | Slovenia | P1-0048 |