Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application.
De Benedetti, S., Di Pisa, F., Fassi, E.M.A., Cretich, M., Musico, A., Frigerio, R., Mussida, A., Bombaci, M., Grifantini, R., Colombo, G., Bolognesi, M., Grande, R., Zanchetta, N., Gismondo, M.R., Mileto, D., Mancon, A., Gourlay, L.J.(2021) Vaccines (Basel) 9
- PubMed: 33915716
- DOI: https://doi.org/10.3390/vaccines9040322
- Primary Citation of Related Structures:
6SSV - PubMed Abstract:
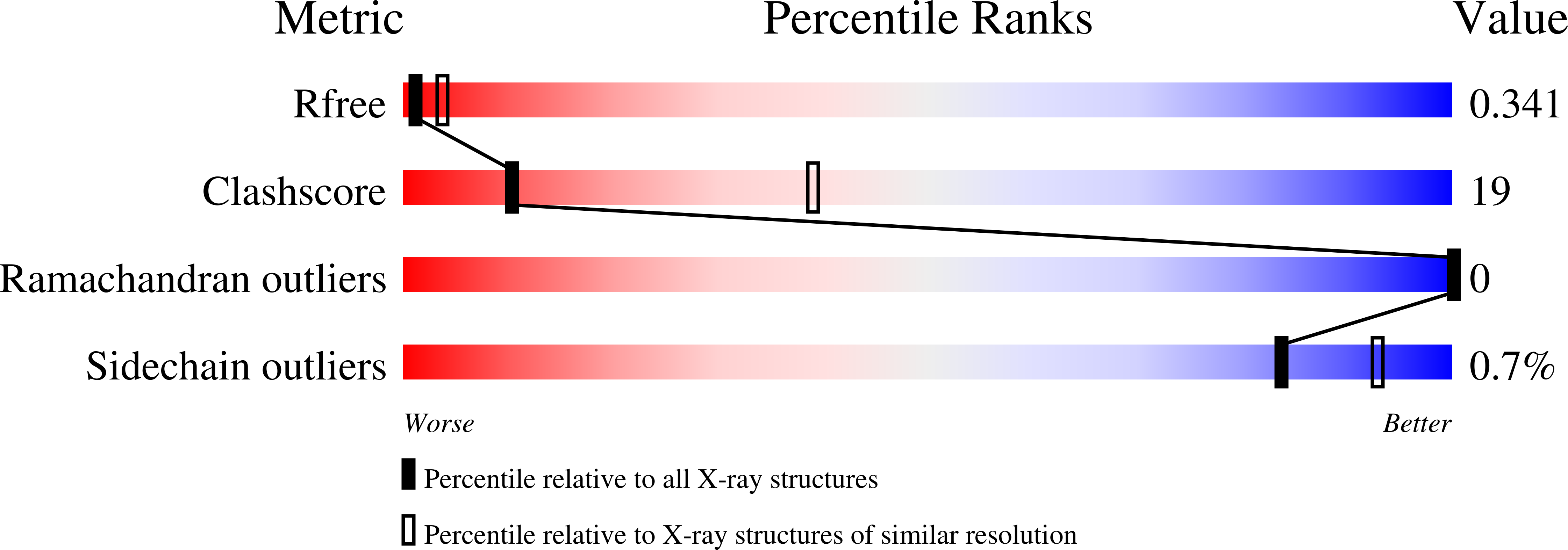
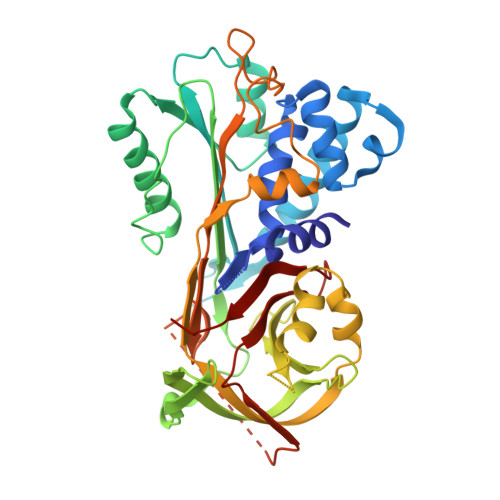
The human parasitic disease Schistosomiasis is caused by the Schistosoma trematode flatworm that infects freshwaters in tropical regions of the world, particularly in Sub-Saharan Africa, South America, and the Far-East. It has also been observed as an emerging disease in Europe, due to increased immigration. In addition to improved therapeutic strategies, it is imperative to develop novel, rapid, and sensitive diagnostic tests that can detect the Schistosoma parasite, allowing timely treatment. Present diagnosis is difficult and involves microscopy-based detection of Schistosoma eggs in the feces. In this context, we present the 3.22 Å resolution crystal structure of the circulating antigen Serine protease inhibitor from S. mansoni (SmSPI), and we describe it as a potential serodiagnostic marker. Moreover, we identify three potential immunoreactive epitopes using in silico-based epitope mapping methods. Here, we confirm effective immune sera reactivity of the recombinant antigen, suggesting the further investigation of the protein and/or its predicted epitopes as serodiagnostic Schistosomiasis biomarkers.
Organizational Affiliation:
Department of Biosciences, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy.