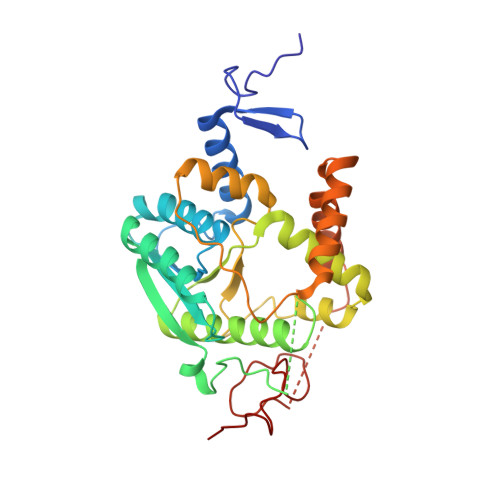
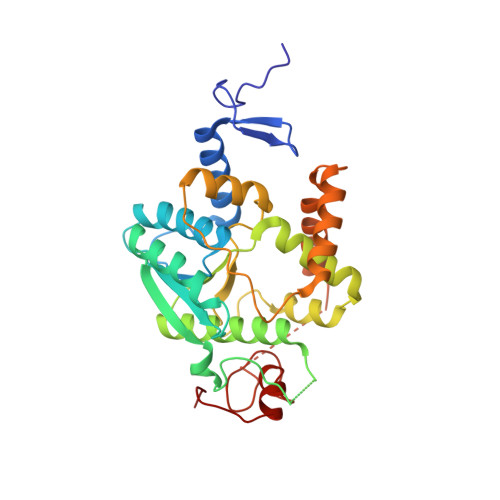
The thiolation of uridine 34 in tRNA, which controls protein translation, depends on a [4Fe-4S] cluster in the archaeum Methanococcus maripaludis.
Bimai, O., Legrand, P., Ravanat, J.L., Touati, N., Zhou, J., He, N., Lenon, M., Barras, F., Fontecave, M., Golinelli-Pimpaneau, B.(2023) Sci Rep 13: 5351-5351
- PubMed: 37005440
- DOI: https://doi.org/10.1038/s41598-023-32423-9
- Primary Citation of Related Structures:
6SCY - PubMed Abstract:
Thiolation of uridine 34 in the anticodon loop of several tRNAs is conserved in the three domains of life and guarantees fidelity of protein translation. U34-tRNA thiolation is catalyzed by a complex of two proteins in the eukaryotic cytosol (named Ctu1/Ctu2 in humans), but by a single NcsA enzyme in archaea. We report here spectroscopic and biochemical experiments showing that NcsA from Methanococcus maripaludis (MmNcsA) is a dimer that binds a [4Fe-4S] cluster, which is required for catalysis. Moreover, the crystal structure of MmNcsA at 2.8 Å resolution shows that the [4Fe-4S] cluster is coordinated by three conserved cysteines only, in each monomer. Extra electron density on the fourth nonprotein-bonded iron most likely locates the binding site for a hydrogenosulfide ligand, in agreement with the [4Fe-4S] cluster being used to bind and activate the sulfur atom of the sulfur donor. Comparison of the crystal structure of MmNcsA with the AlphaFold model of the human Ctu1/Ctu2 complex shows a very close superposition of the catalytic site residues, including the cysteines that coordinate the [4Fe-4S] cluster in MmNcsA. We thus propose that the same mechanism for U34-tRNA thiolation, mediated by a [4Fe-4S]-dependent enzyme, operates in archaea and eukaryotes.
Organizational Affiliation:
Laboratoire de Chimie des Processus Biologiques, Collège de France, CNRS UMR 8229, Sorbonne Université, 11 Place Marcelin Berthelot, 75231, Paris Cedex 05, France.