Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Vogler, M., Karan, R., Renn, D., Vancea, A., Vielberg, M.T., Grotzinger, S.W., DasSarma, P., DasSarma, S., Eppinger, J., Groll, M., Rueping, M.(2020) Front Microbiol 11: 742-742
- PubMed: 32411108
- DOI: https://doi.org/10.3389/fmicb.2020.00742
- Primary Citation of Related Structures:
6SC4 - PubMed Abstract:
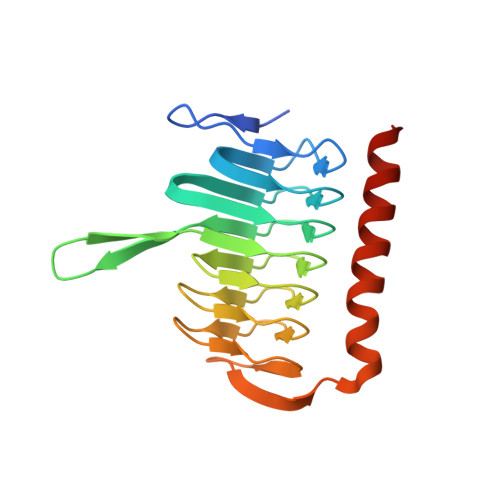
Environments previously thought to be uninhabitable offer a tremendous wealth of unexplored microorganisms and enzymes. In this paper, we present the discovery and characterization of a novel γ-carbonic anhydrase (γ-CA) from the polyextreme Red Sea brine pool Discovery Deep (2141 m depth, 44.8°C, 26.2% salt) by single-cell genome sequencing. The extensive analysis of the selected gene helps demonstrate the potential of this culture-independent method. The enzyme was expressed in the bioengineered haloarchaeon Halobacterium sp. NRC-1 and characterized by X-ray crystallography and mutagenesis. The 2.6 Å crystal structure of the protein shows a trimeric arrangement. Within the γ-CA, several possible structural determinants responsible for the enzyme's salt stability could be highlighted. Moreover, the amino acid composition on the protein surface and the intra- and intermolecular interactions within the protein differ significantly from those of its close homologs. To gain further insights into the catalytic residues of the γ-CA enzyme, we created a library of variants around the active site residues and successfully improved the enzyme activity by 17-fold. As several γ-CAs have been reported without measurable activity, this provides further clues as to critical residues. Our study reveals insights into the halophilic γ-CA activity and its unique adaptations. The study of the polyextremophilic carbonic anhydrase provides a basis for outlining insights into strategies for salt adaptation, yielding enzymes with industrially valuable properties, and the underlying mechanisms of protein evolution.
Organizational Affiliation:
KAUST Catalysis Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia.