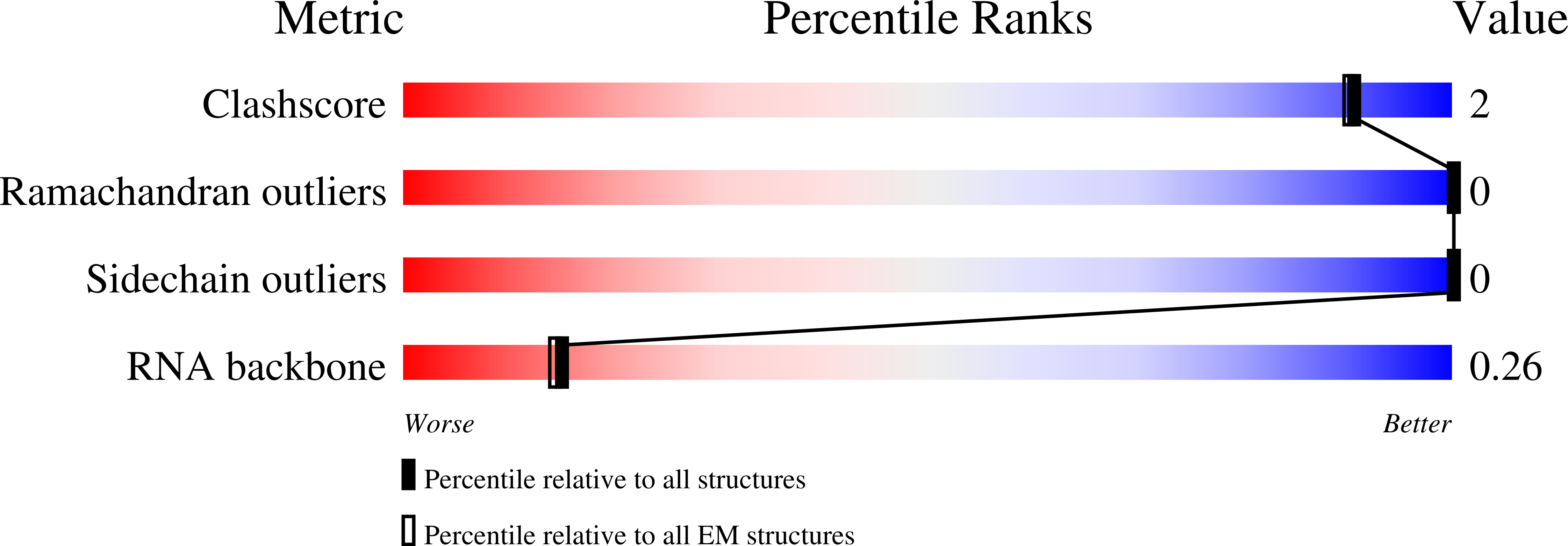Elucidation of the viral disassembly switch of tobacco mosaic virus.
Weis, F., Beckers, M., von der Hocht, I., Sachse, C.(2019) EMBO Rep 20: e48451-e48451
- PubMed: 31535454
- DOI: https://doi.org/10.15252/embr.201948451
- Primary Citation of Related Structures:
6SAE, 6SAG - PubMed Abstract:
Stable capsid structures of viruses protect viral RNA while they also require controlled disassembly for releasing the viral genome in the host cell. A detailed understanding of viral disassembly processes and the involved structural switches is still lacking. This process has been extensively studied using tobacco mosaic virus (TMV), and carboxylate interactions are assumed to play a critical part in this process. Here, we present two cryo-EM structures of the helical TMV assembly at 2.0 and 1.9 Å resolution in conditions of high Ca 2+ concentration at low pH and in water. Based on our atomic models, we identify the conformational details of the disassembly switch mechanism: In high Ca 2+ /acidic pH environment, the virion is stabilized between neighboring subunits through carboxyl groups E95 and E97 in close proximity to a Ca 2+ binding site that is shared between two subunits. Upon increase in pH and lower Ca 2+ levels, mutual repulsion of the E95/E97 pair and Ca 2+ removal destabilize the network of interactions between adjacent subunits at lower radius and release the switch for viral disassembly.
Organizational Affiliation:
Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.