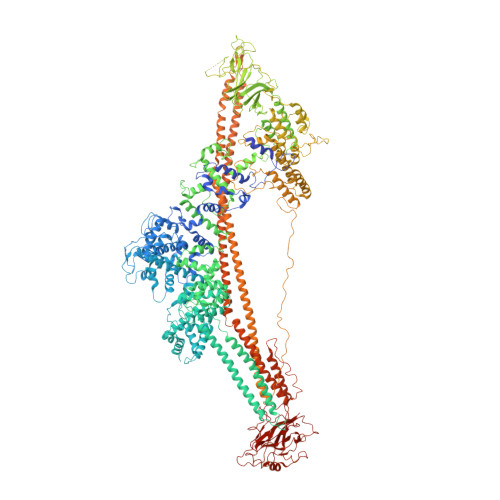
Common architecture of Tc toxins from human and insect pathogenic bacteria.
Leidreiter, F., Roderer, D., Meusch, D., Gatsogiannis, C., Benz, R., Raunser, S.(2019) Sci Adv 5: eaax6497-eaax6497
- PubMed: 31663026
- DOI: https://doi.org/10.1126/sciadv.aax6497
- Primary Citation of Related Structures:
6RW6, 6RW8, 6RW9, 6RWA, 6RWB - PubMed Abstract:
Tc toxins use a syringe-like mechanism to penetrate the membrane and translocate toxic enzymes into the host cytosol. They are composed of three components: TcA, TcB, and TcC. Low-resolution structures of TcAs from different bacteria suggest a considerable difference in their architecture and possibly in their mechanism of action. Here, we present high-resolution structures of five TcAs from insect and human pathogens, which show a similar overall composition and domain organization. Essential structural features, including a trefoil protein knot, are present in all TcAs, suggesting a common mechanism of action. All TcAs form functional pores and can be combined with TcB-TcC subunits from other species to form active chimeric holotoxins. We identified a conserved ionic pair that stabilizes the shell, likely operating as a strong latch that only springs open after destabilization of other regions. Our results provide new insights into the architecture and mechanism of the Tc toxin family.
Organizational Affiliation:
Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Str. 11, 44227 Dortmund, Germany.