Inverse control of Rab proteins byYersiniaADP-ribosyltransferase and glycosyltransferase related to clostridial glucosylating toxins.
Ost, G.S., Wirth, C., Bogdanovic, X., Kao, W.C., Schorch, B., Aktories, P.J.K., Papatheodorou, P., Schwan, C., Schlosser, A., Jank, T., Hunte, C., Aktories, K.(2020) Sci Adv 6: eaaz2094-eaaz2094
- PubMed: 32195351
- DOI: https://doi.org/10.1126/sciadv.aaz2094
- Primary Citation of Related Structures:
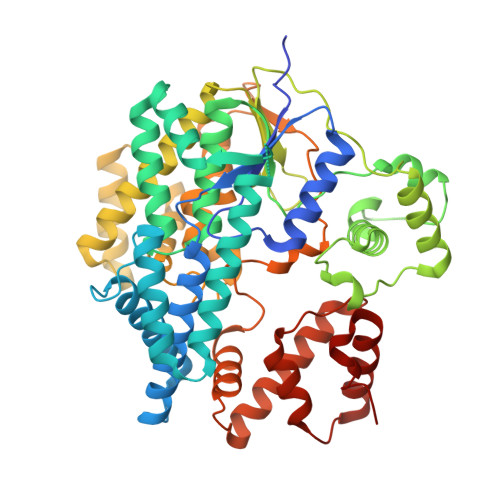
6RTG, 6RTH - PubMed Abstract:
We identified a glucosyltransferase (YGT) and an ADP-ribosyltransferase (YART) in Yersinia mollaretii , highly related to glucosylating toxins from Clostridium difficile , the cause of antibiotics-associated enterocolitis. Both Yersinia toxins consist of an amino-terminal enzyme domain, an autoprotease domain activated by inositol hexakisphosphate, and a carboxyl-terminal translocation domain. YGT N -acetylglucosaminylates Rab5 and Rab31 at Thr 52 and Thr 36 , respectively, thereby inactivating the Rab proteins. YART ADP-ribosylates Rab5 and Rab31 at Gln 79 and Gln 64 , respectively. This activates Rab proteins by inhibiting GTP hydrolysis. We determined the crystal structure of the glycosyltransferase domain of YGT (YGT G ) in the presence and absence of UDP at 1.9- and 3.4-Å resolution, respectively. Thereby, we identified a previously unknown potassium ion-binding site, which explains potassium ion-dependent enhanced glycosyltransferase activity in clostridial and related toxins. Our findings exhibit a novel type of inverse regulation of Rab proteins by toxins and provide new insights into the structure-function relationship of glycosyltransferase toxins.
Organizational Affiliation:
Institut für Experimentelle und Klinische Pharmakologie und Toxikologie, Medizinische Fakultät, Albert-Ludwigs-Universität Freiburg, 79104 Freiburg, Germany.