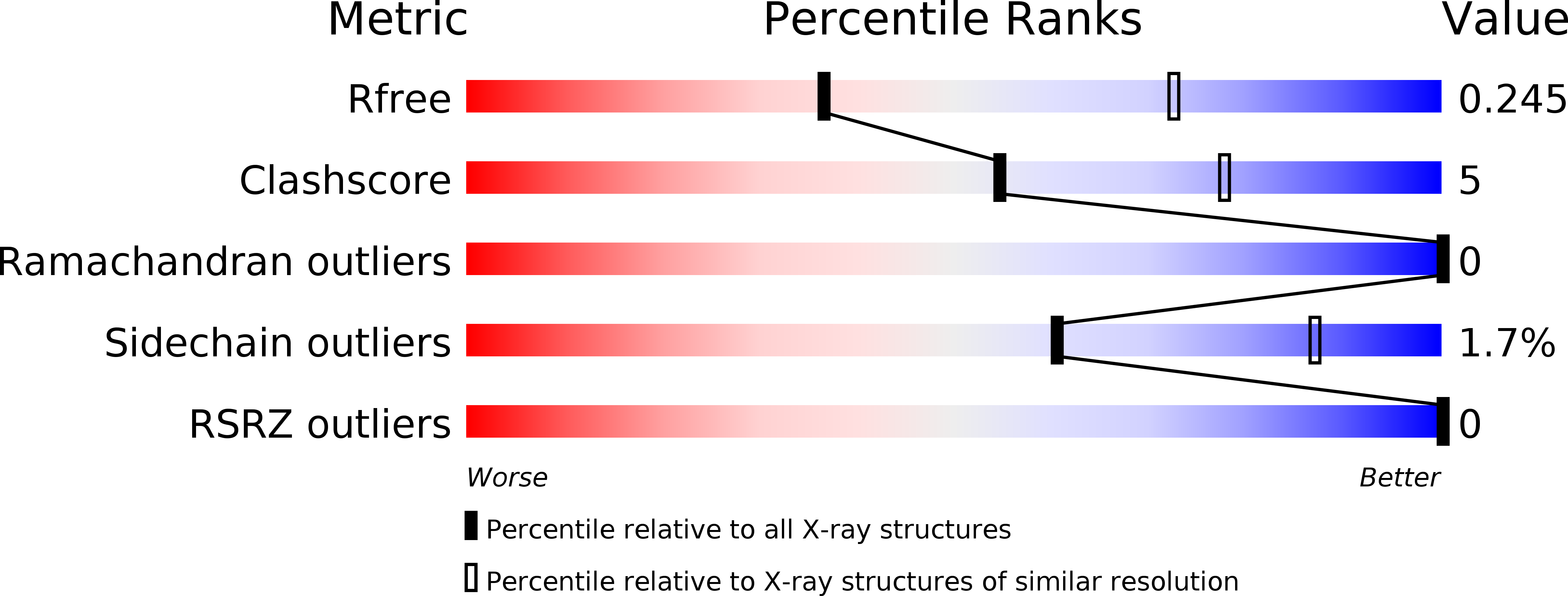
Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Carrique, L., Ballut, L., Shukla, A., Varma, N., Ravi, R., Violot, S., Srinivasan, B., Ganeshappa, U.T., Kulkarni, S., Balaram, H., Aghajari, N.(2020) Nat Commun 11: 3228-3228
- PubMed: 32591529
- DOI: https://doi.org/10.1038/s41467-020-17013-x
- Primary Citation of Related Structures:
6RMD, 6RME, 6RMO, 6RMW, 6RN1, 6RNH - PubMed Abstract:
Plasmodium falciparum (Pf) relies solely on the salvage pathway for its purine nucleotide requirements, making this pathway indispensable to the parasite. Purine nucleotide levels are regulated by anabolic processes and by nucleotidases that hydrolyse these metabolites into nucleosides. Certain apicomplexan parasites, including Pf, have an IMP-specific-nucleotidase 1 (ISN1). Here we show, by comprehensive substrate screening, that PfISN1 catalyzes the dephosphorylation of inosine monophosphate (IMP) and is allosterically activated by ATP. Crystal structures of tetrameric PfISN1 reveal complex rearrangements of domain organization tightly associated with catalysis. Immunofluorescence microscopy and expression of GFP-fused protein indicate cytosolic localization of PfISN1 and expression in asexual and gametocyte stages of the parasite. With earlier evidence on isn1 upregulation in female gametocytes, the structures reported in this study may contribute to initiate the design for possible transmission-blocking agents.
Organizational Affiliation:
Molecular Microbiology and Structural Biochemistry, UMR5086 CNRS-University of Lyon, Lyon, 69007, France.