Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain.
Molla, G., Chaves-Sanjuan, A., Savinelli, A., Nardini, M., Pollegioni, L.(2020) FASEB J 34: 1182-1197
- PubMed: 31914658
- DOI: https://doi.org/10.1096/fj.201901703R
- Primary Citation of Related Structures:
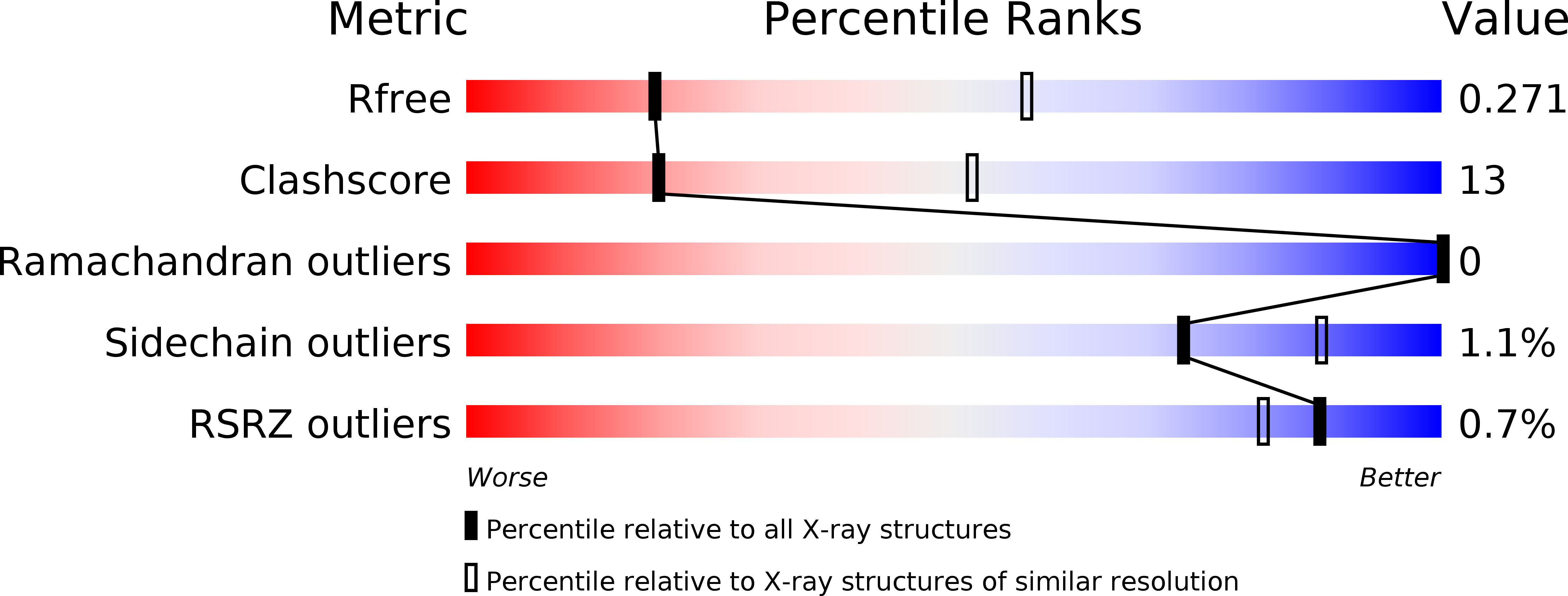
6RKF - PubMed Abstract:
d-Amino acids are the "wrong" enantiomers of amino acids as they are not used in proteins synthesis but evolved in selected functions. On this side, d-aspartate (d-Asp) plays several significant roles in mammals, especially as an agonist of N-methyl-d-aspartate receptors (NMDAR), and is involved in relevant diseases, such as schizophrenia and Alzheimer's disease. In vivo modulation of d-Asp levels represents an intriguing task to cope with such pathological states. As little is known about d-Asp synthesis, the only option for modulating the levels is via degradation, which is due to the flavoenzyme d-aspartate oxidase (DASPO). Here we present the first three-dimensional structure of a DASPO enzyme (from human) which belongs to the d-amino acid oxidase family. Notably, human DASPO differs from human d-amino acid oxidase (attributed to d-serine degradation, the main coagonist of NMDAR) showing peculiar structural features (a specific active site charge distribution), oligomeric state and kinetic mechanism, and a higher FAD affinity and activity. These results provide useful insights into the structure-function relationships of human DASPO: modulating its activity represents now a feasible novel therapeutic target.
Organizational Affiliation:
Dipartimento di Biotecnologie e Scienze della Vita, Università degli studi dell'Insubria, Varese, Italy.