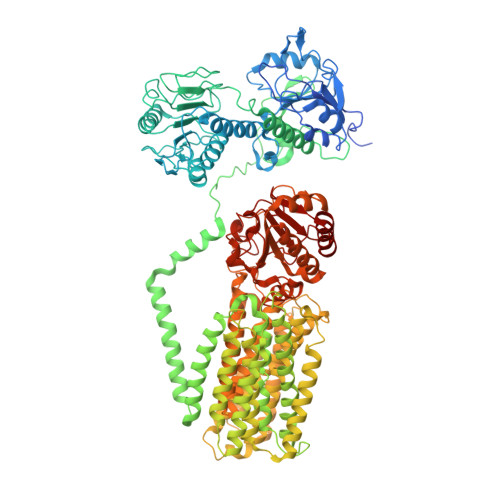
Structure and mechanism of mitochondrial proton-translocating transhydrogenase.
Kampjut, D., Sazanov, L.A.(2019) Nature 573: 291-295
- PubMed: 31462775
- DOI: https://doi.org/10.1038/s41586-019-1519-2
- Primary Citation of Related Structures:
6QTI, 6QUE, 6S59 - PubMed Abstract:
Proton-translocating transhydrogenase (also known as nicotinamide nucleotide transhydrogenase (NNT)) is found in the plasma membranes of bacteria and the inner mitochondrial membranes of eukaryotes. NNT catalyses the transfer of a hydride between NADH and NADP + , coupled to the translocation of one proton across the membrane. Its main physiological function is the generation of NADPH, which is a substrate in anabolic reactions and a regulator of oxidative status; however, NNT may also fine-tune the Krebs cycle 1,2 . NNT deficiency causes familial glucocorticoid deficiency in humans and metabolic abnormalities in mice, similar to those observed in type II diabetes 3,4 . The catalytic mechanism of NNT has been proposed to involve a rotation of around 180° of the entire NADP(H)-binding domain that alternately participates in hydride transfer and proton-channel gating. However, owing to the lack of high-resolution structures of intact NNT, the details of this process remain unclear 5,6 . Here we present the cryo-electron microscopy structure of intact mammalian NNT in different conformational states. We show how the NADP(H)-binding domain opens the proton channel to the opposite sides of the membrane, and we provide structures of these two states. We also describe the catalytically important interfaces and linkers between the membrane and the soluble domains and their roles in nucleotide exchange. These structures enable us to propose a revised mechanism for a coupling process in NNT that is consistent with a large body of previous biochemical work. Our results are relevant to the development of currently unavailable NNT inhibitors, which may have therapeutic potential in ischaemia reperfusion injury, metabolic syndrome and some cancers 7-9 .
Organizational Affiliation:
Institute of Science and Technology Austria, Klosterneuburg, Austria.