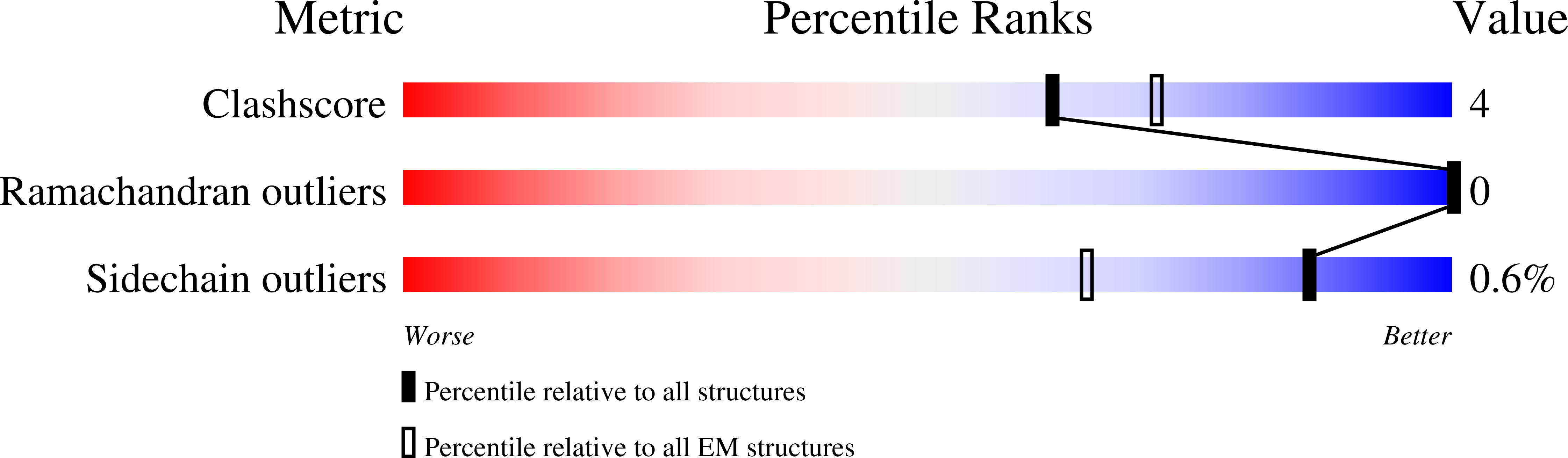Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Cunha, E.S., Chen, X., Sanz-Gaitero, M., Mills, D.J., Luecke, H.(2021) Nat Commun 12: 230-230
- PubMed: 33431861
- DOI: https://doi.org/10.1038/s41467-020-20485-6
- Primary Citation of Related Structures:
6QSU, 6ZJA - PubMed Abstract:
Infection of the human stomach by Helicobacter pylori remains a worldwide problem and greatly contributes to peptic ulcer disease and gastric cancer. Without active intervention approximately 50% of the world population will continue to be infected with this gastric pathogen. Current eradication, called triple therapy, entails a proton-pump inhibitor and two broadband antibiotics, however resistance to either clarithromycin or metronidazole is greater than 25% and rising. Therefore, there is an urgent need for a targeted, high-specificity eradication drug. Gastric infection by H. pylori depends on the expression of a nickel-dependent urease in the cytoplasm of the bacteria. Here, we report the 2.0 Å resolution structure of the 1.1 MDa urease in complex with an inhibitor by cryo-electron microscopy and compare it to a β-mercaptoethanol-inhibited structure at 2.5 Å resolution. The structural information is of sufficient detail to aid in the development of inhibitors with high specificity and affinity.
Organizational Affiliation:
Structural Biology and Drug Discovery Group, Centre for Molecular Medicine Norway, Nordic EMBL Partnership, University of Oslo and Oslo University Hospital, 0318, Oslo, Norway. cunha@uio.no.