The Alkyne Moiety as a Latent Electrophile in Irreversible Covalent Small Molecule Inhibitors of Cathepsin K.
Mons, E., Jansen, I.D.C., Loboda, J., van Doodewaerd, B.R., Hermans, J., Verdoes, M., van Boeckel, C.A.A., van Veelen, P.A., Turk, B., Turk, D., Ovaa, H.(2019) J Am Chem Soc 141: 3507-3514
- PubMed: 30689386
- DOI: https://doi.org/10.1021/jacs.8b11027
- Primary Citation of Related Structures:
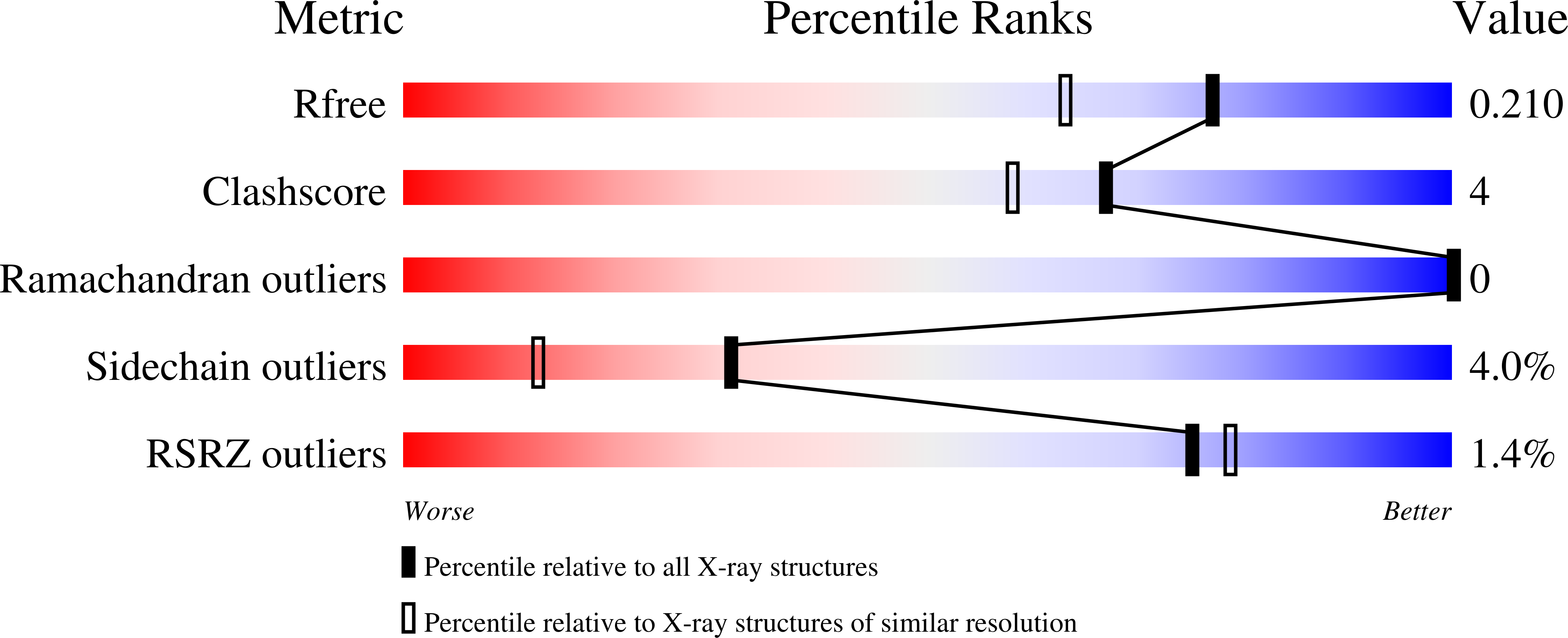
6QBS - PubMed Abstract:
Irreversible covalent inhibitors can have a beneficial pharmacokinetic/pharmacodynamics profile but are still often avoided due to the risk of indiscriminate covalent reactivity and the resulting adverse effects. To overcome this potential liability, we introduced an alkyne moiety as a latent electrophile into small molecule inhibitors of cathepsin K (CatK). Alkyne-based inhibitors do not show indiscriminate thiol reactivity but potently inhibit CatK protease activity by formation of an irreversible covalent bond with the catalytic cysteine residue, confirmed by crystal structure analysis. The rate of covalent bond formation ( k inact ) does not correlate with electrophilicity of the alkyne moiety, indicative of a proximity-driven reactivity. Inhibition of CatK-mediated bone resorption is validated in human osteoclasts. Together, this work illustrates the potential of alkynes as latent electrophiles in small molecule inhibitors, enabling the development of irreversible covalent inhibitors with an improved safety profile.
Organizational Affiliation:
Department of Cell and Chemical Biology, Oncode Institute , Leiden University Medical Center , 2300 RC Leiden , The Netherlands.